1. Introduction
Hyperaccumulator plants are a unique group of species that can accumulate extraordinary concentrations of heavy metals, including cadmium, copper, and zinc. These remarkable plants are essential to phytoremediation strategies because they extract heavy metals from contaminated soils, thereby reducing environmental toxicity and restoring ecosystem function. Their use has become increasingly important for sustainable environmental management [
1,
2,
3,
4,
5,
6,
7,
8].
Over the past decade, research on heavy metal hyperaccumulation and phytoextraction has grown rapidly, reflecting the overall expansion of scientific publishing [
9]. This expansion mirrors the broader trend in scientific publishing, in which researchers face unprecedented challenges in efficiently selecting, structuring, and filtering relevant information from newly published papers. In the context of hyperaccumulation research, the abundance of information creates a critical need for advanced analytical tools that can systematically process scientific literature and summarize parameters such as metal types, concentrations, experimental conditions, and biochemical effects studied across numerous publications. The expansion has produced a large, heterogeneous body of literature reporting on diverse plant species, metal types, concentration ranges, experimental setups, and biochemical responses. While these studies are informative individually, they are difficult to synthesize at a general level using traditional manual approaches.
The emergence of self-attention mechanisms and the subsequent development of large language models (LLMs) have revolutionized our ability to process and extract information from scientific literature [
10,
11,
12,
13,
14]. Recent benchmarks such as SciDaSynth [
15], demonstrate the effective extraction of structured knowledge from the scientific literature. These benchmarks use large multimodal models to interactively process and integrate information from text, tables, and figures. Similarly, in the biomedical field, LLM-powered frameworks have achieved high accuracy in extracting complex data from electronic health records [
16], demonstrating greater reliability and efficiency than conventional manual curation methods. However, general-purpose models still have significant limitations when applied directly to specialized scientific subfields. These limitations include generating fabricated content and hallucinations in specialized domains [
17,
18], difficulties in synthesizing conflicting evidence across multiple studies [
19], and having significant gaps in numerical reasoning and structured scientific data extraction [
20,
21].
Despite these advances, to our knowledge, no specialized LLM tool has been developed to extract information from papers about the hyperaccumulation of heavy metals in plants and phytoextraction. While researchers have made significant attempts to systematize existing knowledge in this field, these efforts have been limited in scope and methodology. For instance, a 2025 study published in Scientific Data [
22] presented a manually curated dataset of phytoremediation parameters for four plant species (sunflower, hemp, castor bean, and bamboo), and extracted 6679 bioconcentration factor observations from 238 papers. However, this labor-intensive approach required manually reviewing 587 studies over seven months, which highlights the significant time and resource constraints of traditional data compilation methods. Similarly, machine learning analysis has been used to predict phytoextraction efficacy factors [
23], incorporating only 173 data points to evaluate various predictive variables. While these efforts provide valuable, structured datasets, researchers currently lack accessible tools for generating custom, complex datasets with broader taxonomic coverage or specific parameters relevant to their research questions. General-purpose LLMs have shown promise in adjacent fields, such as soil health science. There, GPT-4 and specialized tools like GeoGalactica have been used in data processing pipelines [
24]. Similarly, LLM pipelines for processing plant species data have achieved average precision and recall scores exceeding 90% for categorical trait extraction [
25]. Nevertheless, these general applications face limitations, including model overconfidence with limited context, difficulty merging information from distinct sources, and challenges processing numerical data.
Based on our preliminary analysis of scientific data extraction systems, we hypothesize that standard token-level metrics (precision, recall, and F1-score) fail to capture contextual accuracy in scientific knowledge extraction. Specifically, we propose that models with high F1-scores can produce incorrect associations between experimental parameters. This gap in contextual accuracy necessitates a semantic validation layer that evaluates factual consistency rather than term extraction. To address this gap, we present a specialized large language model (LLM) pipeline designed to transform unstructured scientific literature on phytoextraction into a structured knowledge base that complies with the FAIR principles (findability, accessibility, interoperability, and reusability). Our system automatically extracts key parameters, including plant species, metal types, concentrations, experimental setups, and associated biochemical responses, and enables complex querying and trend analysis. Our approach combines multimodal document understanding with rigorous validation, including an “LLM-as-a-Judge” layer, to ensure high recall and factual consistency. This tool enables researchers to generate custom, reproducible datasets at scale, accelerating evidence synthesis and supporting data-driven decisions in environmental science.
2. Materials and Methods
This section outlines our comprehensive methodology for converting unstructured scientific literature on heavy metal hyperaccumulation into structured knowledge. We describe each processing stage, emphasizing the specialized adaptations necessary for accurately handling phytoextraction data. Our pipeline integrates three large language models to balance performance, accessibility, and computational requirements while maintaining scientific integrity through rigorous validation protocols.
2.1. Article Upload and Preprocessing
A custom web server interface allowed users to upload scientific articles in Portable Document Format (PDF). Upon upload, each PDF was processed using the PyMuPDF library to extract individual pages. Then, each page was rendered as a high-resolution PNG image at 300 dots per inch. The Pixmap method, via the Pillow library, was then used to handle the images to ensure consistency in image format.
2.2. Document Conversion to Structured Markdown
The page images were converted into a structured, text-based format to facilitate downstream processing. Google’s Gemini 2.5 Flash was used for this conversion via its public API due to its advanced multimodal capabilities. The model was prompted to generate a comprehensive Markdown representation that preserved the document’s logical structure. The output included:
Hierarchical headings/footers and body text.
Tables converted into Pandas DataFrames (represented as strings and python code).
Mathematical formulas and equations extracted in LaTeX format.
Descriptions of graphical elements (e.g., charts, diagrams); for complex graphs, a textual summary of the key trends and data points was also generated.
References.
2.3. Data Extraction via LLM
The individual Markdown files for each page were concatenated into a single document. This consolidated text was then processed by Qwen3-4B [
26], GPT-OSS-120b [
27] and Gemini 2.5 Pro (via API) to extract specific data entities and convert them into a structured JSON string. For Qwen3-4B and GPT-OSS-120b, inference was performed using llama.cpp with a Vulkan backend. A detailed prompt (available in prompt.md) defined the extraction logic by specifying the target parameters, their expected data types, and the JSON schema.
All models were prompted to calculate the bioconcentration factor (
BCF), which is a fundamental quantitative metric in phytoremediation research.
BCF characterizes a plant’s capacity to accumulate heavy metals from soil media relative to ambient concentrations. The
BCF is formally defined as follows:
where
Cplant represents the concentration of a specific heavy metal in plant tissue, typically reported in mg·kg
−1 of dry weight, and
Csoil denotes the concentration of the same metal species in the corresponding soil or growth substrate under identical experimental conditions. The extraction prompt was engineered to perform multi-step reasoning: First, it identifies paired measurements of plant tissue and soil metal concentrations from the same experimental setup. Second, it verifies unit consistency across measurements. Third, it applies the
BCF formula to compute the ratio. Finally, it preserves traceability to the original source values.
2.4. Validation of Extraction Accuracy
We assessed the correctness of the extracted JSON data using a two-tiered validation approach on a randomly selected sample of processed articles.
Validation against a dataset: The LLM-generated JSON outputs were compared by “AI Judge” (GPT-OSS-120b) against a dataset that was pre-labeled by the authors. The AI judge then calculated standard information retrieval metrics—precision, recall, and F1-score—for each target data field.
Qualitative validation: An independent LLM (GPT-OSS-120b) was used as an “AI judge” to perform a consistency check. This was performed by generating natural language statements based on the JSON content and checking these statements directly against the source text.
2.5. Database Construction
We parsed and ingested validated JSON strings into a MongoDB database. A schema was designed that mapped the nested structure of the JSON data directly to MongoDB documents. This allowed for efficient storage and complex querying.
2.6. Natural Language Query Interface
The system featured a natural language interface for database interrogation. A user’s free-text query was processed by an LLM, which generated a syntactically correct MongoDB query. The generated query was then executed against the database, and the results were sent back to the LLM to formulate a coherent natural language response for the user. All source code and prompts are available in the GitHub repository
https://github.com/KMakrinsky/hyperaccum (accessed on 20 November 2025).
2.7. The Use of GenAI for Manuscript Preparation
Text preparation, including translation, styling, and grammar editing, was performed using GenAI models such as Gemini 2.5 Pro and Qwen3-Max. Gemini 2.5 Pro was also used for code reviews and testing.
3. Results and Discussion
In this section, we comprehensively validate our pipeline’s performance across quantitative metrics and qualitatively assess factual accuracy. We demonstrate how our domain-specific prompt engineering strategy enables the accurate extraction of complex experimental relationships. Our innovative “LLM-as-a-Judge” validation layer reveals the critical limitations of standard evaluation metrics. Beyond technical validation, we present practical applications, such as metal-plant co-occurrence network analysis and the replication of a manually curated dataset that required seven months of expert effort. Together, these results validate our hypothesis that contextual understanding is essential for trustworthy scientific knowledge extraction.
3.1. Overall Pipeline
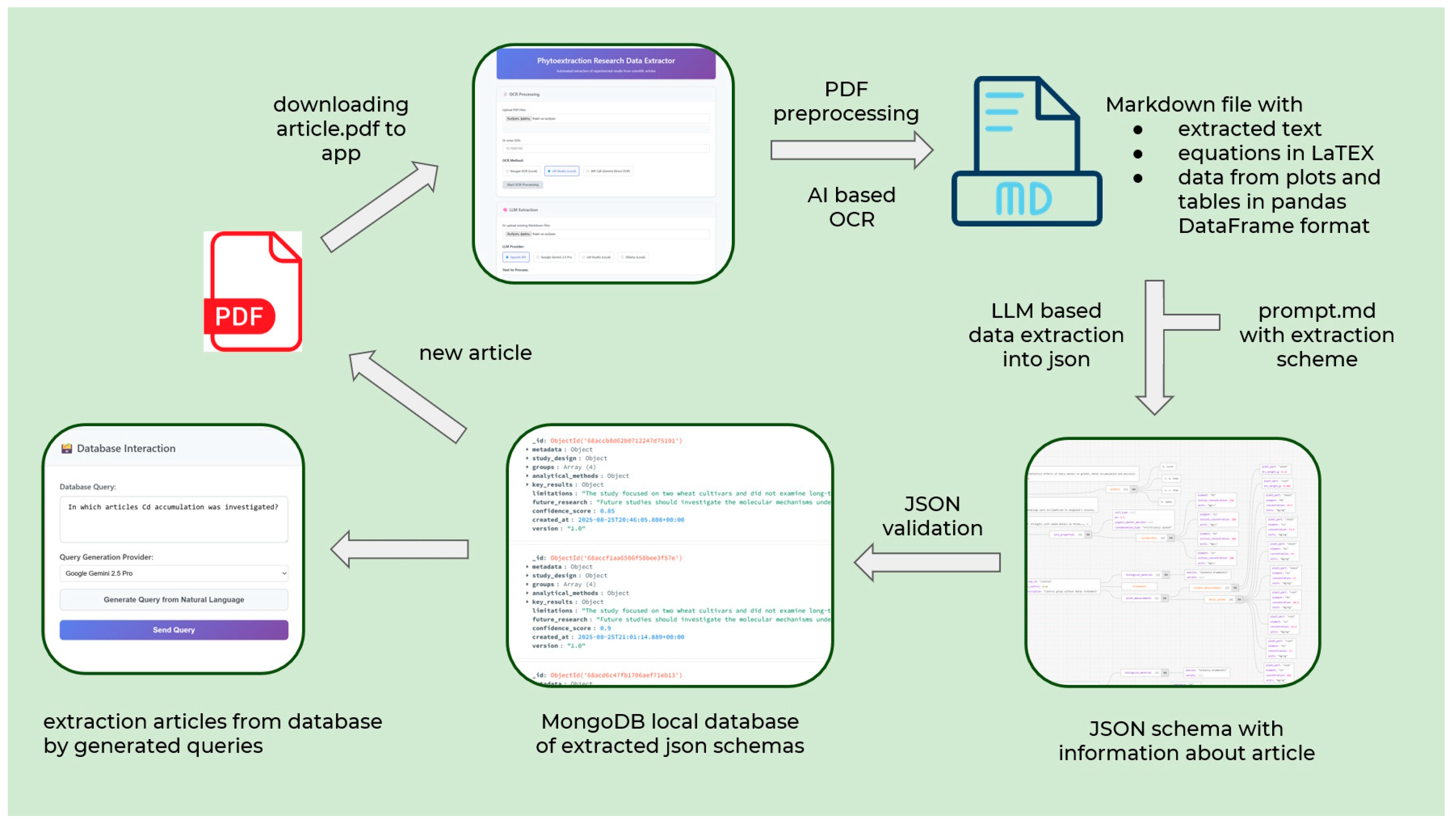
Figure 1 shows our end-to-end process for converting unstructured scientific literature on heavy metal hyperaccumulation into structured knowledge. The system begins with the ingestion of PDF documents and the conversion of high-resolution images, ensuring the preservation of all visual elements, regardless of the complexity of the source formatting.
A key innovation is the multimodal conversion stage, where a multimodal model (Google Gemini 2.5 Flash, in this case) transforms page images into structured Markdown. This process maintains the document’s hierarchy, converts tables to a Pandas-compatible format, preserves mathematical expressions in LaTeX, and generates descriptive summaries of graphical elements. This intermediate representation enables the consistent processing of heterogeneous scientific content prior to extraction.
Knowledge extraction uses specialized LLM prompting with Qwen3-4B, GPT-OSS-120b, and Gemini 2.5 Pro to generate structured JSON outputs containing experimental parameters. The three language models selected for evaluation represent different approaches to scientific text processing. Gemini 2.5 Pro is a state-of-the-art commercial model with advanced multimodal capabilities. GPT-OSS-120b is a large open-source model that strikes a good balance between performance and accessibility. Qwen3-4B illustrates the capabilities of compact models that can be deployed locally with limited computational resources. This selection provides a realistic assessment of the available tools for researchers with different infrastructure constraints and data privacy requirements.
Crucially, our system is built on the foundation of semantic validation for data quality. Rather than relying solely on token-matching metrics, our system uses an “LLM-as-a-Judge” layer to reconstruct natural language statements from extracted JSON subtrees. Then, it verifies the factual correctness of these statements against the source documents. This contextual validation ensures that relationships between entities remain intact. For example, it ensures that specific metal concentrations are linked to the correct plant species under precise experimental conditions. This capability is essential for scientific integrity and cannot be captured by standard precision/recall metrics.
A domain-specific approach was conducted through prompt engineering at every processing stage. Instead of using generic extraction templates, we carefully craft each prompt with hyperaccumulation-specific context and instructions. During the conversion of PDFs to Markdown, the model receives explicit guidance on preserving elements relevant to phytoextraction, such as metal concentration tables and experimental setup descriptions. During extraction, the prompts incorporate specialized knowledge about valid concentration ranges for different metals, unit conversion protocols specific to soil–plant systems, and critical relationships between experimental variables. Even our validation layer incorporates domain expertise about biologically plausible metal accumulation patterns. This end-to-end domain adaptation transforms general-purpose LLMs into specialized scientific assistants that understand the nuanced relationships defining phytoremediation research.
3.2. Quantitative Validation
We benchmarked the performance of three large language models for the task of structured data extraction to evaluate our pipeline’s core: Gemini 2.5 Pro (API-based), Qwen3-4B, and GPT-OSS-120B (both of which are locally hosted).
For each processed article, we created 20 sets of paired validation inputs. The first input was a manually curated CSV record according to our annotation schema. Domain experts created this record using the ground truth. The second input was the machine-generated CSV output from each evaluated LLM. We submitted these paired records to an independent validation model, GPT-OSS-120b, configured as an “AI Judge”, which performed a granular, field-by-field comparison using semantic matching rather than strict string equality. Specifically, the model was prompted to normalize terminology variants (e.g., recognizing “cadmium chloride” and “CdCl2” as equivalent to the target entity “Cd”), and to resolve unit conversions and numerical representations (e.g., recognizing “50 μM” as equivalent to “0.05 mM”).
The judge model automatically computed standard information retrieval metrics—precision, recall, and F1 score—for each target field across all validation samples. This semantic validation approach addresses a critical limitation of conventional token-matching evaluation by preventing the misclassification of technically correct, terminologically variant extractions as false negatives by naive string comparison methods.
Table 1 shows the aggregated performance metrics, revealing significant variations in model performance across parameter categories. This underscores the necessity of context-aware validation for scientific knowledge extraction.
Overall, Gemini 2.5 Pro demonstrated the most robust and balanced performance. It achieved an average F1-score of 0.896 across articles, which was the highest among all models. Qwen-4B followed with an F1 score of 0.871, and GPT-OSS-120b followed with an F1 score of 0.854. These results suggest that while all models can perform the extraction task well, larger, proprietary models currently have an advantage in terms of overall accuracy and reliability.
A deeper analysis of specific data fields revealed a clear pattern: all models excel at extracting simple, well-defined entities. For instance, performance was exceptionally high in identifying plant species (plants F1) and heavy metals (metals F1). Gemini 2.5-Pro and Qwen-3-4B achieved perfect F1 scores of 1.00 for plant identification, while Gemini achieved a nearly perfect score of 0.975 for metals. This success can be attributed to the fact that these entities are typically discrete nouns or standardized terms (e.g., “
Arabidopsis thaliana”, “Cd”, and “Zinc”), which are analogous to standard named entity recognition (NER) tasks in which modern LLMs are known to perform well. [
28].
In contrast, extracting more complex, descriptive information posed the greatest challenge to all models. The methods and groups yielded the lowest F1 scores consistently. This discrepancy underscores a critical point: lower performance reflects not only model limitations but also the inherent complexity and lack of standardization in scientific writing. Unlike discrete entities, experimental methods are often described in a narrative style. The distribution of crucial details across multiple paragraphs, supplementary materials, and figure legends is a well-documented obstacle in automated information extraction and a known contributor to the “reproducibility crisis” in science [
29].
Similarly, the concept of an “experimental group” is often ambiguous. Authors may describe treatments without explicitly labeling them as distinct groups. Control conditions are often implied, such as “plants grown under standard conditions”, rather than explicitly defined. This requires the model to make complex inferences and synthesize fragmented information across the text. This task is so challenging that even human annotators tasked with extracting such complex information often achieve only moderate inter-annotator agreement. Studies on creating scientific corpora have consistently shown that agreement scores are highest for simple entities and decrease sharply for complex, relational, or descriptive information, such as experimental protocols [
30,
31]. Therefore, the models’ struggles in these categories highlight their difficulty in understanding linguistic complexity and the conceptual ambiguity within scientific discourse. This remains a challenging task even for human experts.
Interestingly, the two locally run open-source models exhibited distinct behavioral profiles. GPT-OSS-120b demonstrated a high-precision, low-recall strategy, particularly for identifying experimental groups (group precision of 0.905 versus group recall of 0.860). This suggests a more “cautious” model that extracts data only when highly confident, potentially resulting in the omission of relevant information. Conversely, Qwen-3-4B showed higher recall (group recall of 0.876) but lower precision (group precision of 0.773). This indicates a more “eager” approach that successfully captures more relevant data points, albeit with some incorrect ones included. The trade-off between precision and recall is critical when selecting a model for a specific application, depending on whether avoiding false positives or false negatives is more important.
3.3. Qualitative Validation
Although F1 scores confirm that a model can identify the correct words in a text, they do not guarantee that the model understands the critical relationships between those words. For a scientific database, this distinction is important. For instance, a paper may state that Arabidopsis halleri was treated with 50 µM of Cd and Thlaspi rotundifolium with 100 µM of Cd. A model with limited contextual understanding could correctly identify both species and concentrations, which would boost its F1 score. However, it could then create a JSON entry that incorrectly links A. halleri to the 100 µM concentration.
To detect such critical errors, we implemented the “AI judge” validation procedure, which aligns with the “LLM-as-a-Judge” paradigm [
32]. A growing body of research comparing its performance against human judgments across diverse tasks supports the credibility and reliability of the LLM-as-a-Judge approach [
33]. In information retrieval, the approach has been validated for scalable relevance judgment [
34] and for the comprehensive evaluation of Retrieval-Augmented Generation (RAG) systems, assessing parameters such as relevance, accuracy, and coherence [
35]. Together, these studies substantiate the LLM-as-a-Judge methodology as a robust, scalable alternative to traditional evaluation metrics. The schema of our validation approach is presented in
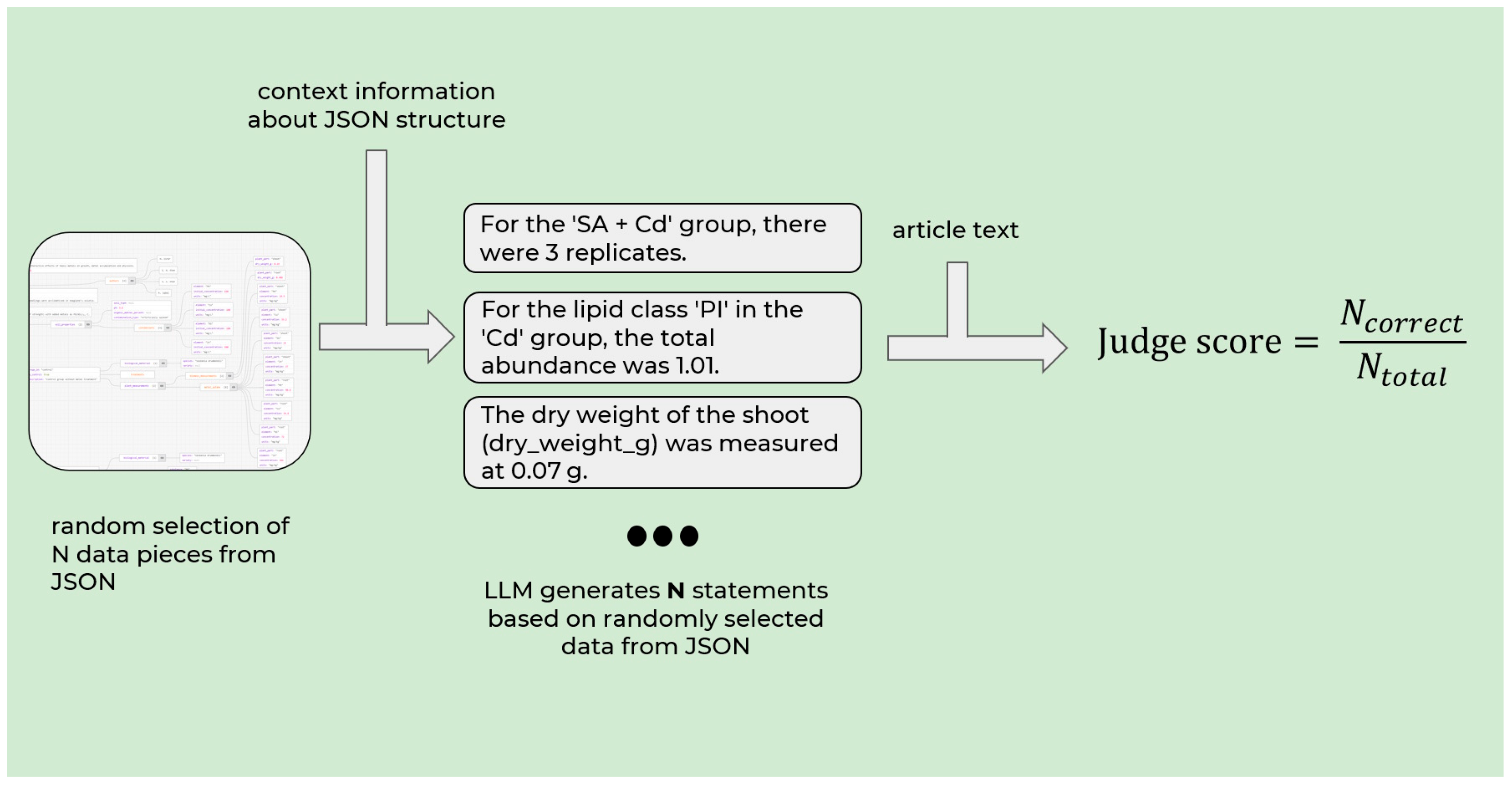
Figure 2.
In this approach, the validation model (GPT-OSS-120b) was given not only the extracted JSON fragment, but also the full hierarchical context leading to each individual parameter. Specifically, for each extracted field (e.g., metal concentration linked to a plant species), the system reconstructed the complete JSON subtree—from the root document object through experimental groups, plant entries, and metal records—to preserve the semantic relationships encoded during extraction. The judge LLM then used this contextual subtree to generate a factual statement in natural language (e.g., “In the study,
Arabidopsis halleri was exposed to 50 µM of cadmium under hydroponic conditions”). Then, the same LLM was prompted to independently verify this statement against the original, full-text Markdown representation of the source article without access to the extracted JSON. Only if the claim could be unambiguously confirmed from the source text was the extraction deemed factually correct. This two-step process—context-aware claim generation and source-grounded verification—ensures that entities are correctly identified and that their relational integrity is preserved. This mitigates hallucinated or misaligned data entries that standard token-level metrics fail to capture. The resulting scores are presented in
Table 2.
The dramatic decrease in Qwen3-4b’s performance—from an F1 score of 0.871 to a qualitative accuracy of 46.7%—underscores its vulnerability to contextual errors. Its high-recall strategy allows it to identify keywords relevant to phytoextraction studies, such as concentrations, chemical compounds, and analytical methods. However, it often fails to correctly assemble the full experimental picture. This failure mode is a form of “hallucination”, which, in this scientific context, manifests as generating non-existent experimental results. This type of error is problematic because it can mislead researchers who query the database, causing them to draw incorrect conclusions about a plant’s tolerance or accumulation capacity under specific conditions.
In contrast, the stronger performance of Gemini 2.5 Pro and GPT-OSS-120b suggests a greater ability for “compositional reasoning”, or the capacity to correctly link specific parameters, such as metal concentration, soil pH, and exposure duration, to the corresponding plant species or experimental group.
This dual-validation approach confirms that a qualitative, fact-checking validation layer is essential, not just beneficial, to build a trustworthy knowledge base in a specialized field such as heavy metal hyperaccumulation. This validation layer ensures that the curated data accurately represents scientific findings and is not just a collection of extracted terms.
3.4. Usage Strategies
Our pipeline’s utility lies in its ability to transform unstructured documents into a searchable knowledge base, which goes beyond validating the extraction process. This enables researchers to synthesize information and identify large-scale trends in the field of hyper-accumulation, rather than analyzing individual papers. The natural language query interface provides access to this synthesized knowledge.
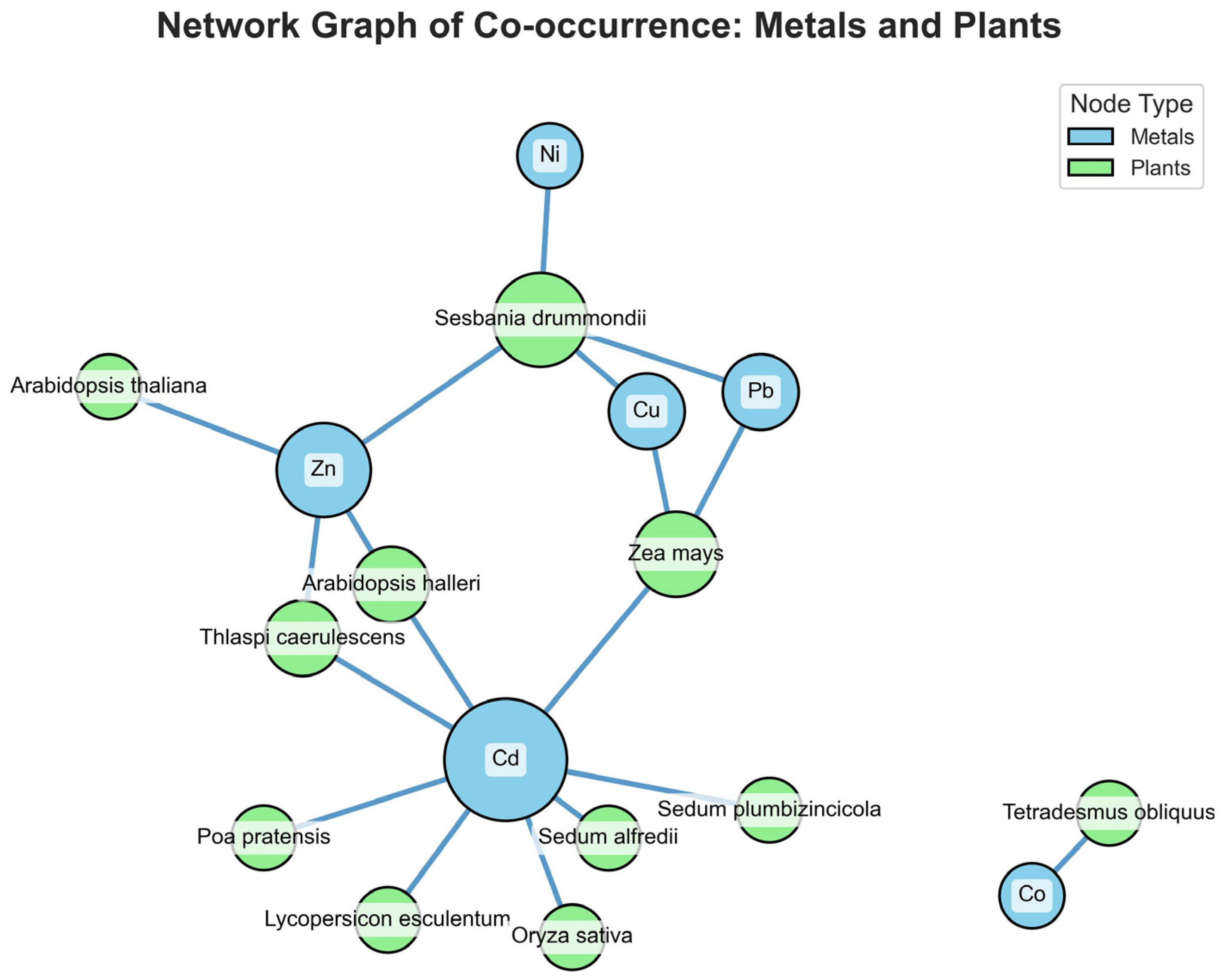
To demonstrate this capability, we analyzed a sample corpus of fifteen articles and generated a co-occurrence network graph of the metals and plants mentioned in the processed articles (
Figure 3). In this graph, nodes represent either a plant species or a heavy metal. An edge between two nodes indicates that the corresponding entities were studied together in at least one paper within our sample corpus.
The resulting visualization reveals key trends within the sampled literature. Cadmium emerges as a central hub with the highest degree of connectivity, linking to a wide array of plant species, including Sedum alfredii, Arabidopsis halleri, and Oryza sativa. This indicates that Cd toxicity and accumulation are the most extensively studied topics in our dataset, reflecting Cd status as a widespread and hazardous environmental pollutant. Zinc is also a significant research focus, sharing connections with key model organisms, such as A. halleri and T. caerulescens, which are known co-accumulators of Cd and Zn.
Additionally, the graph identifies plant species with unique properties. For instance, Sesbania drummondii is connected to multiple metals (Pb, Ni, and Cu), suggesting that it could be the subject of multi-metal phytoremediation research. Conversely, the isolated pair of Tetrademum obliquum (an alga) and cobalt suggests a niche area of research that is disconnected from the main body of terrestrial plant studies in our corpus. Such visualizations provide researchers entering the field with a map of the research landscape, highlighting dominant themes, key model systems, and potential knowledge gaps.
3.5. Replicating a Human-Curated Dataset via LLM Reasoning
To validate the accuracy of our pipeline against established scientific standards and demonstrate its flexibility, we chose to reproduce the manually curated dataset referenced [
22]. This dataset is a gold standard in phytoextraction research, but it took seven months of expert effort to compile. Thanks to our system’s schema-agnostic architecture, this replication required minimal effort. Only the extraction prompts needed modification, and no changes were necessary to the underlying pipeline infrastructure.
Specifically, we created a specialized prompt for Gemini 2.5 Pro, available as prompt_dataset.md in our repository, that precisely mirrored the schema of the target dataset. This prompt instructed the model to extract parameters that matched the exact column structure of the published tables; calculate derived metrics, such as the bioconcentration factor (BCF), on the fly; and consolidate values across similar experimental conditions (e.g., plant components, fertilizer treatments) according to meta-analysis conventions in phytoextraction research. We changed the expected JSON output schema so that it would mirror the flat table format of the target dataset instead of our default hierarchical structure. We provided explicit definitions for each parameter, including its scientific meaning, acceptable value ranges, units of measurement, and data type specifications. This parameter-by-parameter guidance ensured that the extracted data was fully structurally compatible with the original dataset.
This approach allowed for a direct, one-to-one comparison between our automated outputs and the manually curated reference data. For sample articles from the dataset, we averaged
BCF values for each plant-metal combination across comparable experimental conditions. This allowed us to quantitatively assess extraction accuracy while adhering to standard practices for consolidating data in environmental meta-analyses. To establish robust performance metrics, we randomly selected ten articles from the original dataset. For this sample, our pipeline achieved an average relative error of 8.21% for
BCF values. The remarkable fidelity of this replication, achieved solely through prompt adaptation, demonstrates our pipeline’s capacity to rapidly generate custom datasets matching any required structure. This transforms a process that traditionally required months of manual work into one that can be completed in minutes per article.
Table 3 provides a representative example of results for several selected articles from the original dataset.
When we applied our pipeline to an existing phytoextraction dataset, we observed that the LLM effectively replicated existing human-curated datasets with high fidelity. For example, in study [
36], our LLM pipeline captured element-specific accumulation patterns across five distinct hemp varieties (
Zenit,
Diana,
Denise,
Armanca,
Silvana). We observed only minor deviations (4–12%) in directly comparable
BCF values. Similar deviations were observed in [
37], which we attribute to the challenges of processing visually complex tables and figures. In these cases, the multimodal conversion stage occasionally misaligned data points or misinterpreted formatting conventions.
For articles with simpler data presentation formats and fewer experimental points, such as [
38], our approach achieved near-perfect replication of
BCF values, with negligible relative differences (often 0–1%). This variation in performance correlates with the visual complexity of the source data presentations rather than the underlying scientific content. These results demonstrate that, while our pipeline maintains high accuracy with complex layouts, it achieves optimal performance when source documents present data in clear, well-structured formats.
The most important aspect of this approach, beyond the fidelity of the results, is the noticeable acceleration of the entire data curation process. Manually curating a single data-rich scientific article, as was done for the original dataset, can require several hours of a domain expert’s focused effort, including reading, identifying relevant values, performing calculations, and entering data into a structured format. Our automated pipeline completes the same task (from PDF upload to generation of structured, calculated JSON) in a matter of minutes.
4. Conclusions
Our study confirms the hypothesis that standard extraction metrics do not guarantee factual reliability in scientific knowledge extraction. This reveals a gap in contextual accuracy and challenges current evaluation practices in scientific AI and shows that semantic validation through natural language reasoning is necessary for creating reliable knowledge bases. We demonstrate that, although modern LLMs can achieve high extraction performance on discrete entities, their ability to preserve factual relationships—such as linking a metal concentration to the correct plant species—varies significantly. A dual-validation strategy that integrates standard metrics with a qualitative “LLM-as-a-Judge” layer reveals that high F1 scores do not guarantee contextual accuracy, particularly for narrative-rich scientific content, such as experimental protocols. This underscores the necessity of semantic verification in scientific knowledge extraction.
Crucially, our pipeline is prompt-driven and schema-agnostic. By modifying the instruction prompt, we successfully replicated the exact structure of a manually curated benchmark dataset. This allowed us to achieve comparable accuracy in minutes per article versus hours of expert effort. Since all prompts and code are publicly accessible, researchers can quickly adapt the pipeline to their own data schemas. This enables the on-demand generation of custom, FAIR-compliant datasets. The resulting knowledge base supports trend analysis, gap identification, and evidence synthesis—essential capabilities for data-driven environmental science. As an open, reproducible, and flexible framework, this tool offers a scalable solution for phytoremediation research and any other field requiring structured knowledge extraction from scientific literature.