Exploring New Horizons: fNIRS and Machine Learning in Understanding PostCOVID-19
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. fNIRS Data Acquisition
2.3. fNIRS Data Pre-Processing
2.4. Feature Extraction and Classification
2.5. Metrics for Evaluation
3. Results
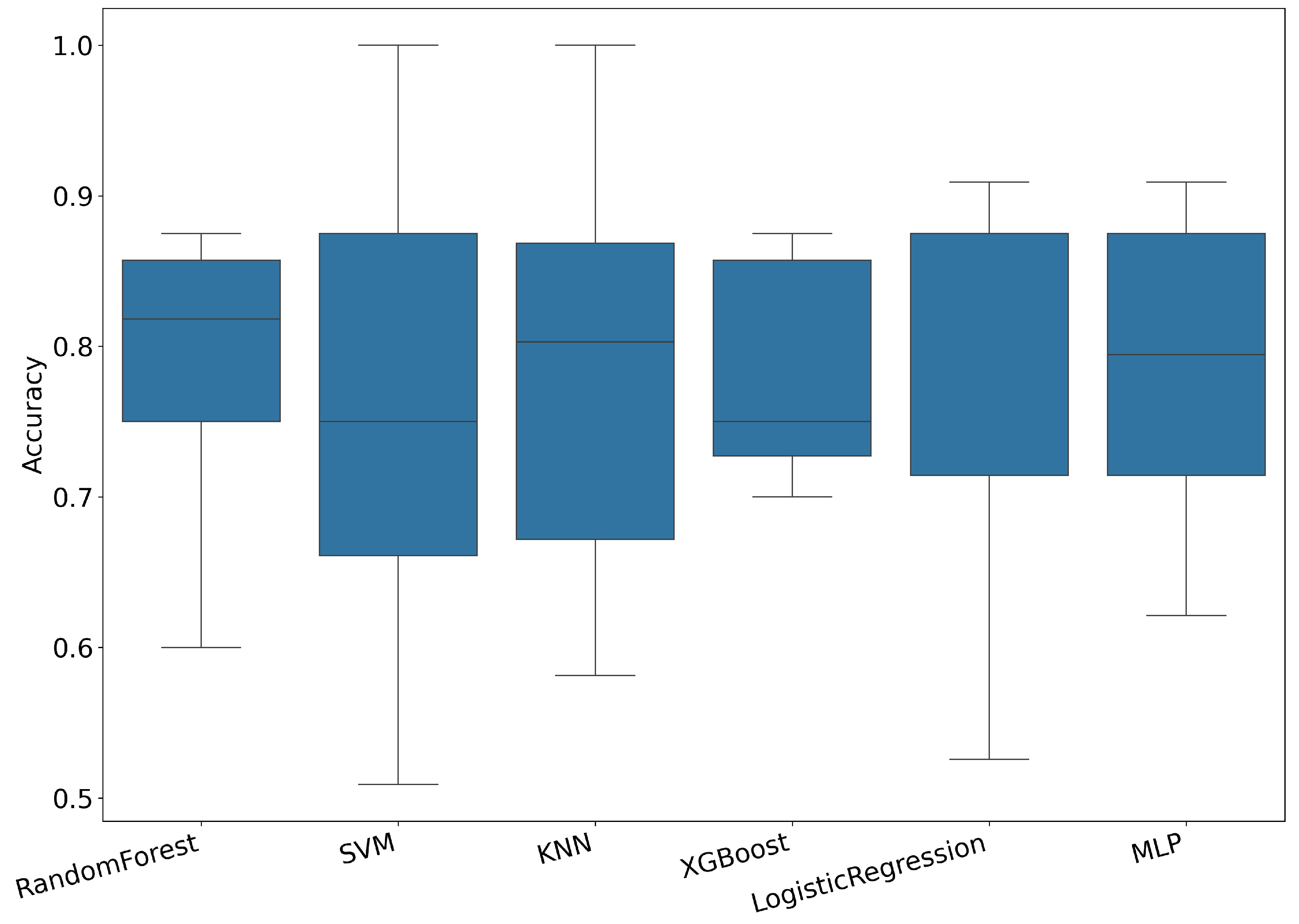
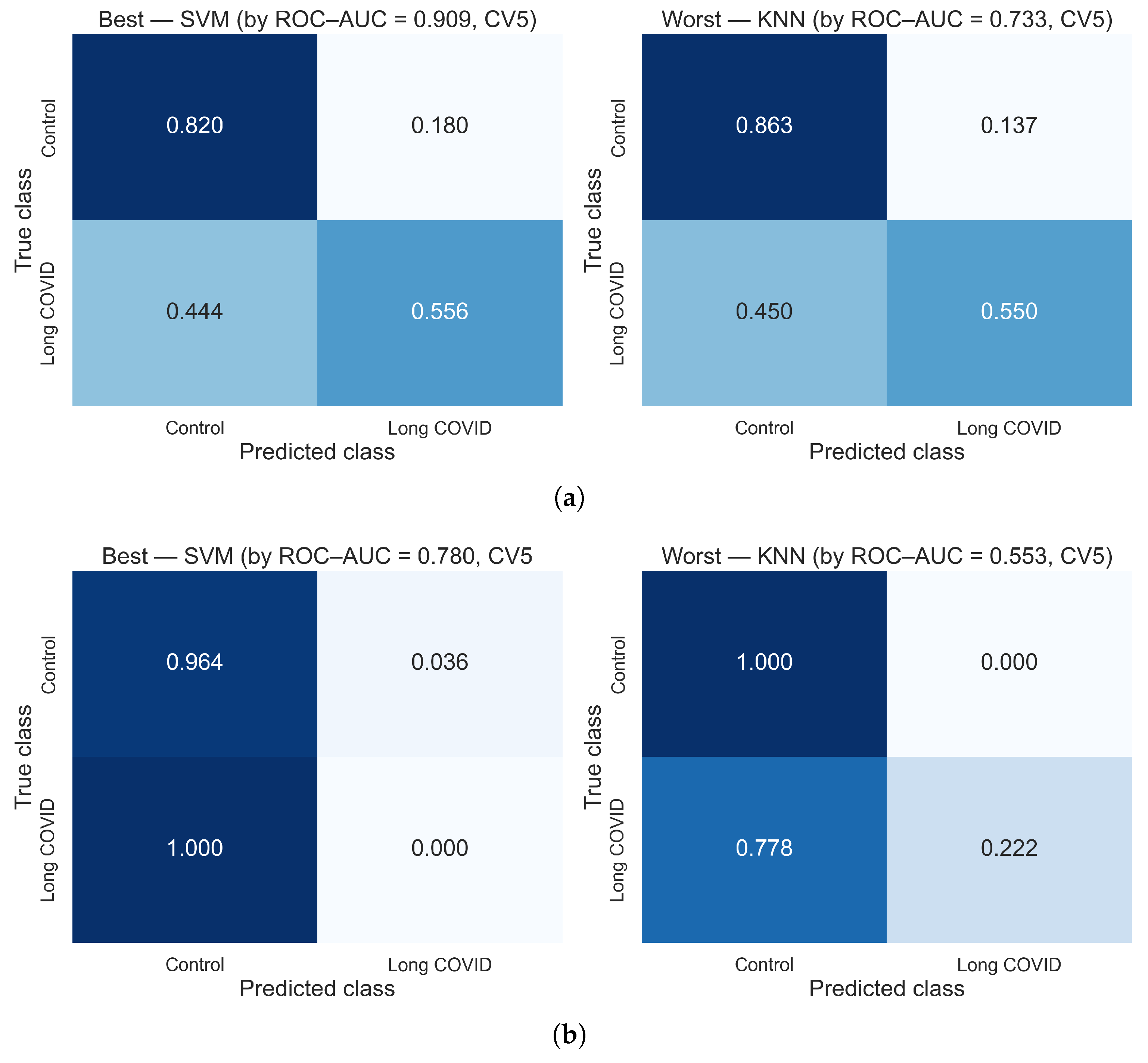
3.1. Results Using Hybrid Features
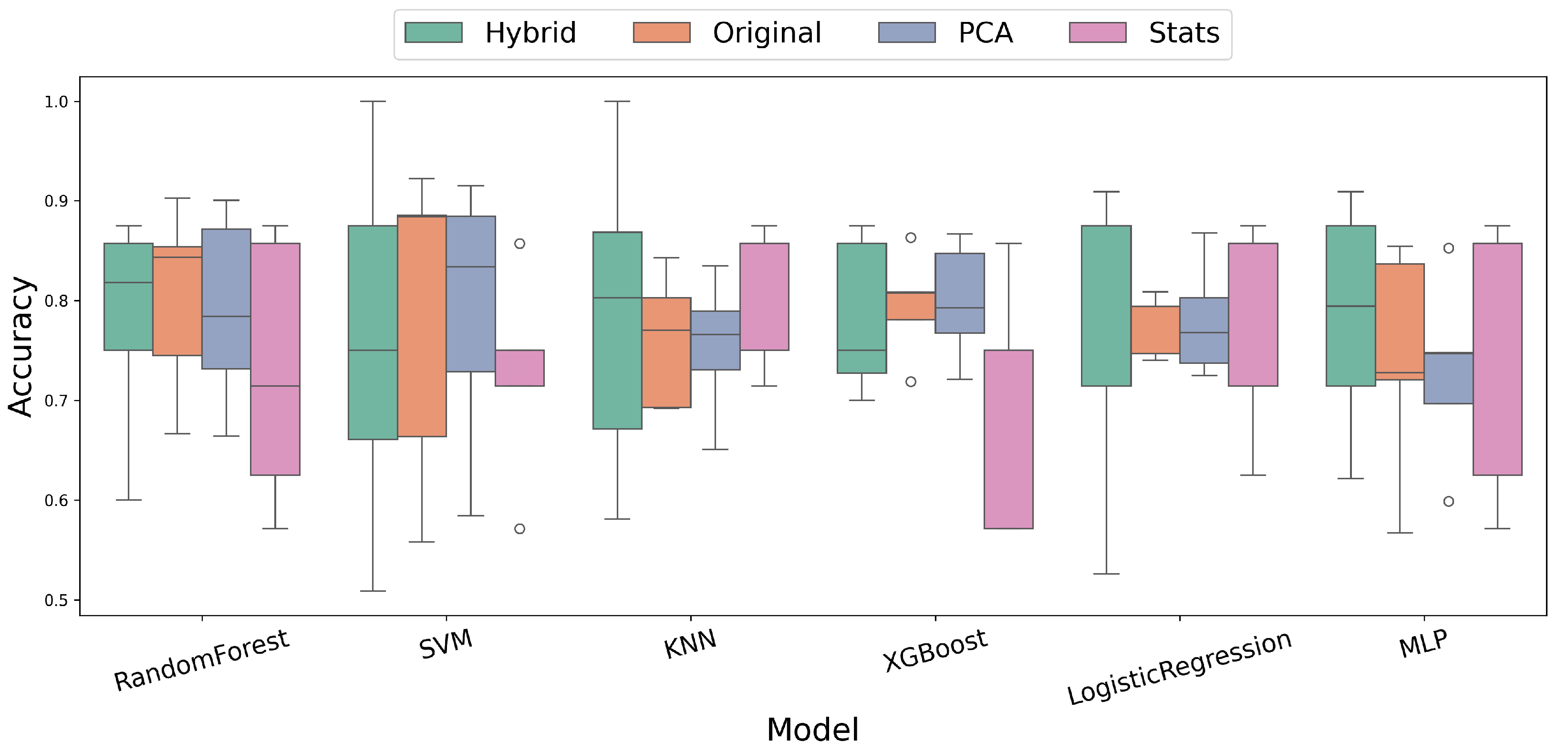
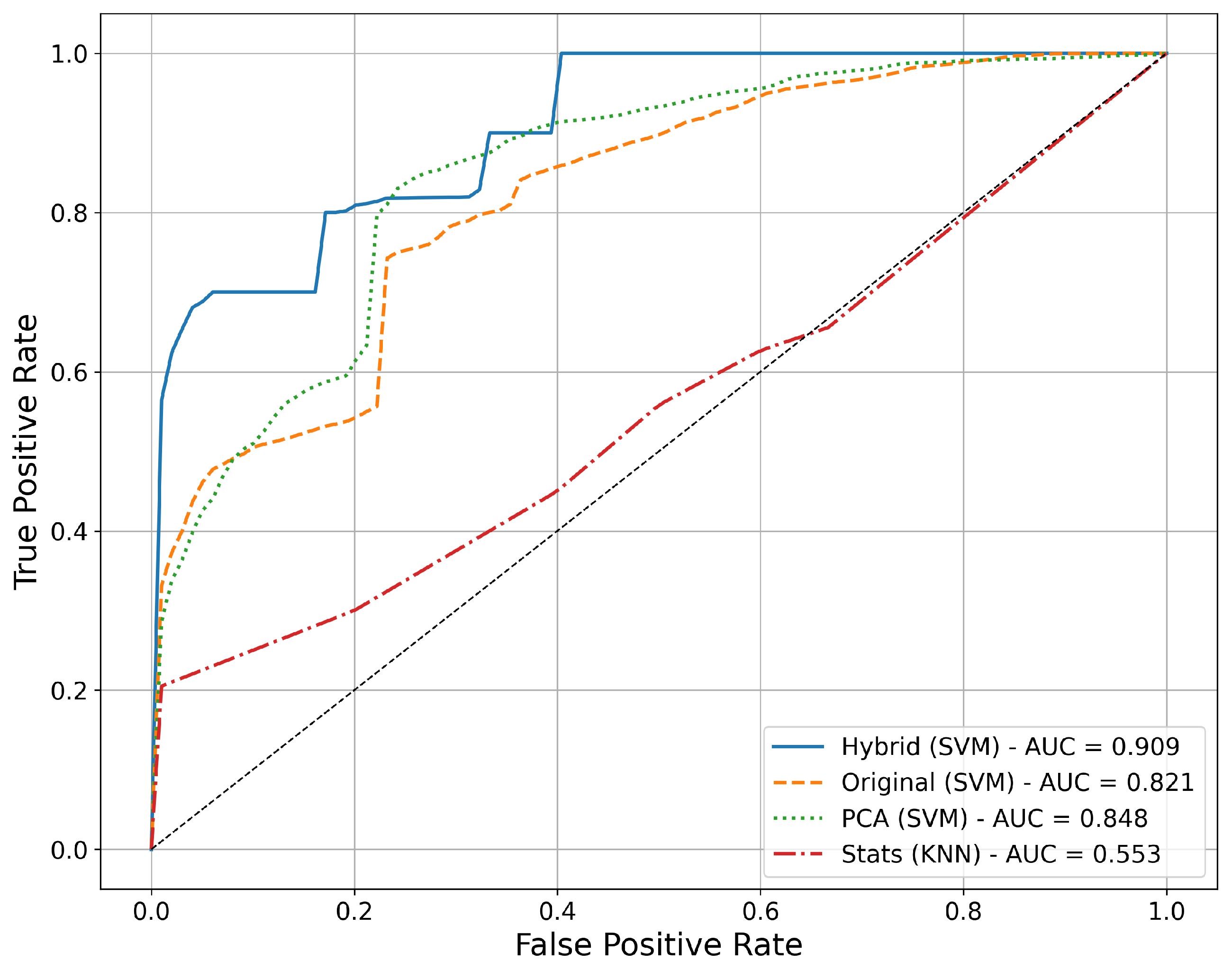
3.2. Comparative Analysis of Feature Extraction Methods
3.3. Feature-Level Interpretation and Informative Channels
3.3.1. Random Forest Feature Importance (Supervised)
3.3.2. PCA Loadings (Unsupervised)
3.3.3. Convergence of Supervised and Unsupervised Evidence
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Post COVID-19 Condition. 2023. Available online: https://www.paho.org/es/temas/coronavirus/brote-enfermedad-por-coronavirus-covid-19/condicion-post-covid-19 (accessed on 28 September 2024).
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Long COVID Statistics. 2024. Available online: https://www.cdc.gov/covid/?CDC_AAref_Val=https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects.html (accessed on 28 September 2024).
- Seo, J.W.; Kim, S.E.; Kim, Y.; Kim, E.J.; Kim, T.; Kim, T.; Lee, S.H.; Lee, E.; Lee, J.; Seo, Y.B.; et al. Updated Clinical Practice Guidelines for the Diagnosis and Management of Long COVID. Infect. Chemother. 2024, 56, 122–157. [Google Scholar] [CrossRef] [PubMed]
- Mina, Y.; Enose-Akahata, Y.; Hammoud, D.A.; Videckis, A.J.; Narpala, S.R.; O’Connell, S.E.; Carroll, R.; Lin, B.C.; McMahan, C.C.; Nair, G.; et al. Deep phenotyping of neurologic postacute sequelae of SARS-CoV-2 infection. Neurol. Neuroimmunol. Neuroinflamm. 2023, 10, e200097. [Google Scholar] [CrossRef] [PubMed]
- Serrano del Pueblo, V.M.; Serrano-Heras, G.; Romero Sánchez, C.M.; Piqueras Landete, P.; Rojas-Bartolome, L.; Feria, I.; Morris, R.G.M.; Strange, B.; Mansilla, F.; Zhang, L.; et al. Brain and cognitive changes in patients with long COVID compared with infection-recovered control subjects. Brain 2024, 147, 3611–3623. [Google Scholar] [CrossRef]
- Greene, C.; Connolly, R.; Brennan, D.; Laffan, A.; O’Keeffe, E.; Zaporojan, L.; O’Callaghan, J.; Thomson, B.; Connolly, E.; Argue, R.; et al. Blood–brain barrier disruption and sustained systemic inflammation in individuals with long COVID-associated cognitive impairment. Nat. Neurosci. 2024, 27, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Sim, S.; Yang, J.; Kim, M. Multivariate prediction of long COVID headache in adolescents using gray matter structural MRI features. Front. Hum. Neurosci. 2023, 17, 1202103. [Google Scholar] [CrossRef]
- Bhattacharya, B.S.; Mitchell, E.G.; Shetty, G.; Delong, G.; Munia, T.T.K.; Tariq, A.A. A Long COVID Risk Predictor Focused on Clinical Workflow Integration. medRxiv 2023. [Google Scholar] [CrossRef]
- Epsi, N.J.; Lindholm, D.A.; Ganesan, A.; Lalani, T.; Smith, A.G.; Mody, R.; Jones, M.; Bazan, S.; Colombo, R.E.; Colombo, C. Precision phenotyping of “long COVID” through machine learning. Open Forum Infect. Dis. 2022, 9, ofac492.907. [Google Scholar] [CrossRef]
- Chen, C.; Li, R.; Shen, H.; Xia, L. Long Short-Term Memory Based Framework for Longitudinal Assessment of COVID-19 Using CT Imaging and Laboratory Data. IEEE Access 2022, 10, 55533–55545. [Google Scholar] [CrossRef]
- Barnden, L.; Thapaliya, K.; Eaton-Fitch, N.; Barth, M.; Marshall-Gradisnik, S. Altered brain connectivity in long COVID during cognitive exertion: A pilot study. Front. Neurosci. 2023, 17, 1182607. [Google Scholar] [CrossRef]
- García-González, L.; Marti-Sarrias, A.; Puertas, M.C.; Bayón-Gil, Á.; Resa-Infante, P.; Martinez-Picado, J.; Navarro, A.; Acosta, S. Understanding the neurological implications of acute and long COVID using brain organoids. Dis. Model. Mech. 2023, 16, dmm049902. [Google Scholar] [CrossRef]
- Matias-Guiu, J.A.; Díez-Cirarda, M. Are there cognitive and neuroimaging signatures in long COVID? Brain Commun. 2023, 5, fcad189. [Google Scholar] [CrossRef]
- Kim, J.; Young, G.S. Neuroimaging of COVID-19. Semin. Neurol. 2023, 43, 205–218. [Google Scholar] [CrossRef]
- Ayaz, H.; Baker, W.B.; Blaney, G.; Boas, D.A.; Bortfeld, H.; Brady, K.; Brake, J.; Brigadoi, S.; Buckley, E.M.; Carp, S.A.; et al. Optical imaging and spectroscopy for the study of the human brain: Status report. Neurophotonics 2022, 9, S24001. [Google Scholar] [CrossRef]
- Lodha, N.; McCarthy, S. Do fine motor impairments constitute a key feature of mild cognitive impairment? A systematic review and meta-analysis. Alzheimer’s Dement. 2021, 17, e056433. [Google Scholar] [CrossRef]
- de Paula, J.J.; Albuquerque, M.R.; Lage, G.M.; Bicalho, M.A.; Romano-Silva, M.A.; Malloy-Diniz, L.F. Impairment of fine motor dexterity in mild cognitive impairment and Alzheimer’s disease dementia: Association with activities of daily living. Rev. Bras. Psiquiatr. 2016, 38, 235–238. [Google Scholar] [CrossRef]
- Hayward, W.; Buch, E.R.; Norato, G.; Iwane, F.; Dash, D.; Salamanca-Girón, R.F.; Bartrum, E.; Walitt, B.; Nath, A.; Cohen, L.G. Procedural motor memory deficits in patients with long-COVID. Neurology 2024, 102, e208073. [Google Scholar] [CrossRef]
- Sanal-Hayes, N.E.M.; Hayes, L.D.; Mclaughlin, M.; Berry, E.C.J.; Sculthorpe, N.F. People with long COVID and ME/CFS exhibit similarly impaired dexterity and bimanual coordination: A case-case-control study. Am. J. Med. 2025, 138, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Sun, C.W.; Lee, P.L.; Lee, H.C.; Yang, C.; Jiang, C.P.; Tong, Y.P.; Yeh, T.C.; Hsieh, J.C. Study of photon migration with various source-detector separations in near-infrared spectroscopic brain imaging based on three-dimensional Monte Carlo modeling. Opt. Express 2005, 13, 8339–8348. [Google Scholar] [CrossRef] [PubMed]
- Brigadoi, S.; Cooper, R.J. How short is short? Optimum source–detector distance for short-separation channels in functional near-infrared spectroscopy. Neurophotonics 2015, 2, 025005. [Google Scholar] [CrossRef]
- Tachtsidis, I.; Scholkmann, F. False positives and false negatives in functional near-infrared spectroscopy: Issues, challenges, and the way forward. Neurophotonics 2016, 3, 031405. [Google Scholar] [CrossRef]
- Yeung, M.K. An optical window into brain function in children and adolescents: A systematic review of functional near-infrared spectroscopy studies. Neuroimage 2021, 227, 117672. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Fox, S.; Blasi, A.; Elwell, C.E. Illuminating the developing brain: The past, present and future of functional near infrared spectroscopy. Neurosci. Biobehav. Rev. 2010, 34, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Huppert, T.J.; Diamond, S.G.; Franceschini, M.A.; Boas, D.A. HomER: A review of time-series analysis methods for near-infrared spectroscopy of the brain. Appl. Opt. 2009, 48, D280–D298. [Google Scholar] [CrossRef]
- Scholkmann, F.; Spichtig, S.; Muehlemann, T.; Wolf, M. How to detect and reduce movement artifacts in near-infrared imaging using moving standard deviation and spline interpolation. Physiol. Meas. 2010, 31, 649–662. [Google Scholar] [CrossRef]
- Selb, J.J.; Yücel, M.A.; Phillip, D.; Schytz, H.W.; Iversen, H.K.; Vangel, M.; Ashina, M.; Boas, D.A. Effect of motion artifacts and their correction on near-infrared spectroscopy oscillation data: A study in healthy subjects and stroke patients. J. Biomed. Opt. 2015, 20, 056011. [Google Scholar] [CrossRef] [PubMed]
- Novi, S.L.; Roberts, E.; Spagnuolo, D.; Spilsbury, B.M.; Price, D.C.; Imbalzano, C.A.; Forero, E.; Tellis, G.M.; Tellis, C.M.; Mesquita, R.C. Functional near-infrared spectroscopy for speech protocols: Characterization of motion artifacts and guidelines for improving data analysis. Neurophotonics 2020, 7, 015001. [Google Scholar] [CrossRef]
- Di Rosa, E.; Brigadoi, S.; Cutini, S.; Tarantino, V.; Dell’Acqua, R.; Mapelli, D.; Braver, T.S.; Vallesi, A. Reward motivation and neurostimulation interact to improve working memory performance in healthy older adults: A simultaneous tDCS-fNIRS study. Neuroimage 2019, 202, 116062. [Google Scholar] [CrossRef]
- Pereira, J.; Direito, B.; Lührs, M.; Castelo-Branco, M.; Sousa, T. Multimodal assessment of the spatial correspondence between fNIRS and fMRI hemodynamic responses in motor tasks. Sci. Rep. 2023, 13, 2244. [Google Scholar] [CrossRef]
- Yücel, M.A.; Lühmann, A.V.; Scholkmann, F.; Gervain, J.; Dan, I.; Ayaz, H.; Boas, D.; Cooper, R.J.; Culver, J.; Elwell, C.E.; et al. Best practices for fNIRS publications. Neurophotonics 2021, 8, 012101. [Google Scholar] [CrossRef]
- Delpy, D.T.; Cope, M.; van der Zee, P.; Arridge, S.; Wray, S.; Wyatt, J. Estimation of optical pathlength through tissue from direct time of flight measurement. Phys. Med. Biol. 1988, 33, 1433–1442. [Google Scholar] [CrossRef]
- Gagnon, L.; Cooper, R.J.; Yücel, M.A.; Perdue, K.L.; Greve, D.N.; Boas, D.A. Short separation channel location impacts the performance of short channel regression in NIRS. Neuroimage 2012, 59, 2518–2528. [Google Scholar] [CrossRef]
- Saager, R.B.; Berger, A.J. Direct characterization and removal of interfering absorption trends in two-layer turbid media. J. Opt. Soc. Am. A 2005, 22, 1874–1882. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.; Yu, M.; Yu, W.; Frederick, B.d.; Tong, Y. Systemic low-frequency oscillations observed in the periphery of healthy human subjects. J. Biomed. Opt. 2018, 23, 057001. [Google Scholar] [CrossRef]
- Chang, L.; Ryan, M.C.; Liang, H.; Zhang, X.; Cunningham, E.; Wang, J.; Wilson, E.; Herskovits, E.H.; Kottilil, S.; Ernst, T.M. Changes in brain activation patterns during working memory tasks in people with post-COVID condition and persistent neuropsychiatric symptoms. Neurology 2023, 100, e2409–e2423. [Google Scholar] [CrossRef]
- Tanashyan, M.M.; Kuznetsova, P.I.; Morozova, S.N.; Annushkin, V.A.; Raskurazhev, A.A. Neuroimaging correlates of post-COVID-19 symptoms: A functional MRI approach. Diagnostics 2024, 14, 2180. [Google Scholar] [CrossRef]
- Strens, L.H.A.; Fogelson, N.; Shanahan, P.; Rothwell, J.C.; Brown, P. The ipsilateral human motor cortex can functionally compensate for acute contralateral motor cortex dysfunction. Curr. Biol. 2003, 13, 1201–1205. [Google Scholar] [CrossRef] [PubMed]
- Clemente, L.; La Rocca, M.; Quaranta, N.; Iannuzzi, L.; Vecchio, E.; Brunetti, A.; Gentile, E.; Dibattista, M.; Lobasso, S.; Bevilacqua, V.; et al. Prefrontal dysfunction in post-COVID-19 hyposmia: An EEG/fNIRS study. Front. Hum. Neurosci. 2023, 17, 1240831. [Google Scholar] [CrossRef] [PubMed]
- Benerradi, J.; Clos, J.; Landowska, A.; Valstar, M.F.; Wilson, M.L. Benchmarking framework for machine learning classification from fNIRS data. Front. Neuroergonom. 2023, 4, 994969. [Google Scholar] [CrossRef]
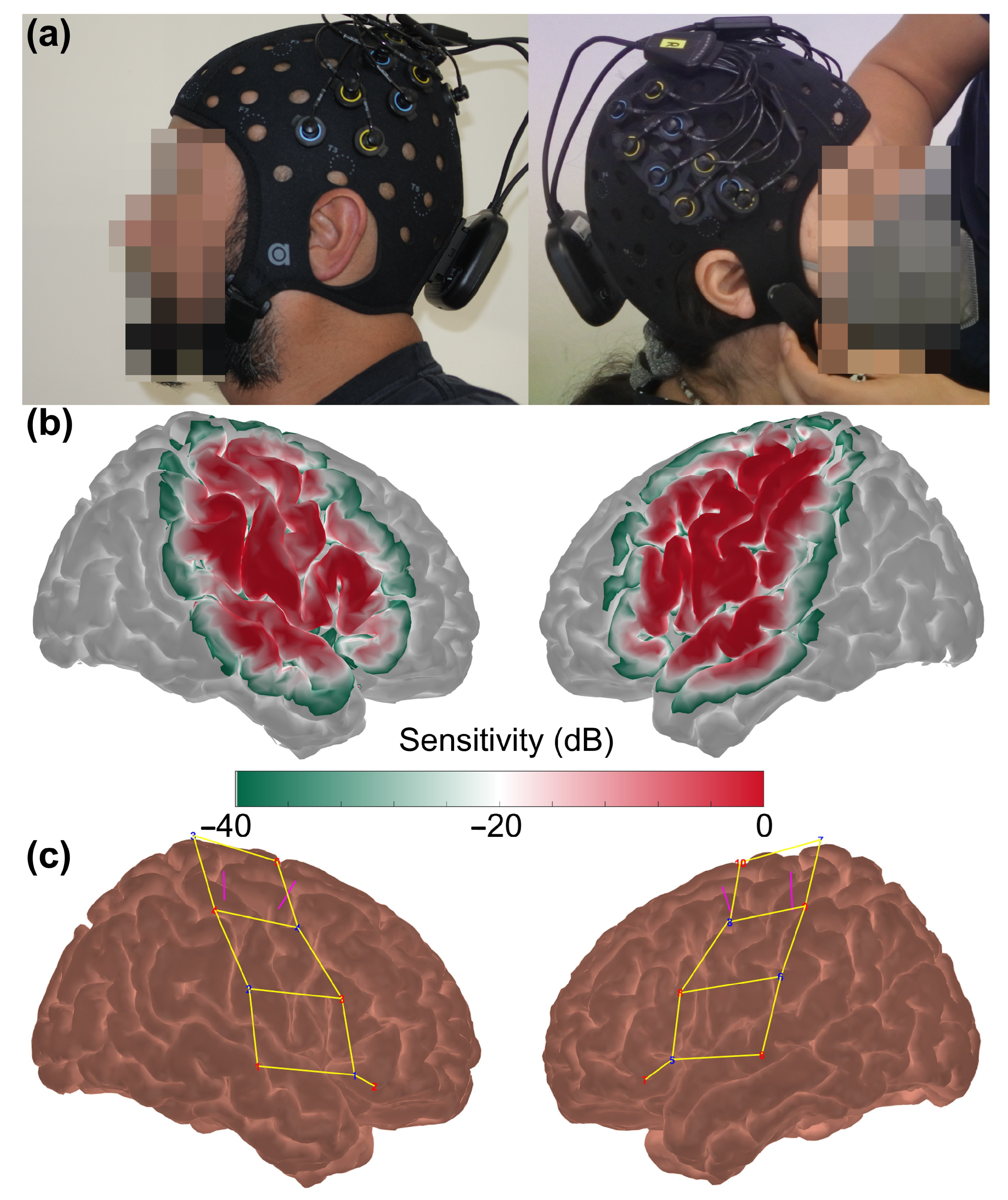


| Parameter | PostCOVID | Control | p-Value |
|---|---|---|---|
| Age (years) | 47.0 [42.0, 61.0] | 61.0 [40.0, 69.5] | 0.3755 1 |
| BMI (kg/m2) | 24.0 [23.4, 30.7] | 24.3 [23.0, 26.1] | 0.6580 1 |
| MoCA | 24.0 [21.5, 26.5] | 27.0 [25.5, 29.0] | 0.02971 |
| Days post-infection | 184.0 [63.0, 187.8] | 125.0 [96.5, 173.0] | 0.7603 1 |
| Number of infections | 2.0 [1.0, 2.0] | 1.0 [1.0, 1.5] | 0.4356 1 |
| SO2 (%) | 90.0 [88.0, 95.0] | 94.0 [92.0, 95.5] | 0.3540 1 |
| FVC (%) | 78.0 [67.8, 81.5] | 91.0 [67.0, 95.5] | 0.0786 1 |
| FEV1 (%) | 76.0 [63.8, 77.0] | 95.0 [71.5, 97.5] | 0.0538 1 |
| PEF (%) | 78.0 [68.0, 85.5] | 97.5 [90.5, 98.5] | 0.0550 1 |
| PBD/FEV1 | 12.0 [2.2, 13.5] | 2.5 [1.0, 10.5] | 0.4291 1 |
| Number of vaccines | 2.0 [1.0, 2.0] | 2.0 [2.0, 3.5] | 0.0956 1 |
| Proportion of males (%) | 33.3% | 57.0% | 0.2691 2 |
| Proportion of hospitalizations (%) | 20.0% | 0.0% | 0.4706 2 |
| Proportion of participants who required oxygen (%) | 20.0% | 0.0% | 0.4706 2 |
| Proportion of hypoxic cases (%) | 55.6% | 25.0% | 0.3348 2 |
| Model | Accuracy | |||||
|---|---|---|---|---|---|---|
| Random Forest | 78.0 | 40.0 | 91.1 | 44.0 | 85.2 | 83.9 |
| SVM | 75.9 | 60.0 | 83.6 | 51.8 | 87.8 | 90.9 |
| KNN | 78.5 | 59.5 | 87.1 | 57.4 | 87.3 | 73.3 |
| XGBoost | 78.2 | 40.0 | 91.1 | 45.0 | 84.8 | 77.8 |
| Logistic Regression | 78.0 | 50.0 | 89.5 | 63.5 | 86.6 | 85.1 |
| MLP | 78.3 | 43.6 | 91.6 | 64.2 | 85.1 | 80.2 |
| Model | |||||
|---|---|---|---|---|---|
| Performance Using Hybrid Features (Original + Statistical) | |||||
| Logistic Regression | 78.0 | 85.1 | 80.8 | 65.7 | 0.453 |
| KNN | 78.5 | 73.3 | 51.5 | 68.9 | 0.443 |
| Random Forest | 78.0 | 83.9 | 81.2 | 59.7 | 0.327 |
| SVM | 75.9 | 90.9 | 81.2 | 65.7 | 0.429 |
| MLP | 78.3 | 80.2 | 73.5 | 63.0 | 0.409 |
| XGBoost | 78.2 | 77.8 | 57.2 | 60.6 | 0.312 |
| Performance Using Original Time-Series | |||||
| Logistic Regression | 77.7 | 64.4 | 52.6 | 64.3 | 0.322 |
| KNN | 76.0 | 63.0 | 31.9 | 59.9 | 0.245 |
| Random Forest | 80.2 | 80.4 | 62.3 | 68.1 | 0.471 |
| SVM | 78.3 | 82.1 | 63.3 | 71.0 | 0.492 |
| MLP | 74.1 | 57.7 | 41.6 | 62.4 | 0.275 |
| XGBoost | 79.6 | 71.9 | 45.3 | 59.6 | 0.281 |
| Performance Using PCA Features (95% Variance) | |||||
| Logistic Regression | 78.0 | 69.4 | 60.6 | 66.2 | 0.363 |
| KNN | 75.4 | 62.9 | 32.0 | 58.8 | 0.222 |
| Random Forest | 79.0 | 79.2 | 55.1 | 67.7 | 0.432 |
| SVM | 78.9 | 84.8 | 62.9 | 70.5 | 0.490 |
| MLP | 72.9 | 55.3 | 41.4 | 58.7 | 0.208 |
| XGBoost | 79.9 | 76.6 | 57.1 | 63.0 | 0.357 |
| Performance Using Statistical Features | |||||
| Logistic Regression | 78.6 | 66.3 | 57.5 | 61.6 | 0.276 |
| KNN | 81.1 | 55.3 | 43.1 | 57.8 | 0.260 |
| Random Forest | 78.2 | 75.7 | 69.2 | 64.7 | 0.339 |
| SVM | 72.9 | 78.0 | 70.7 | 42.0 | −0.052 |
| MLP | 78.6 | 73.0 | 61.2 | 61.6 | 0.276 |
| XGBoost | 70.0 | 75.7 | 60.0 | 45.7 | −0.037 |
| Random Forest (Top–10) | PCA (Top–10) | |||
|---|---|---|---|---|
| Rank | Feature | Rank | Feature | |
| 1 | min_2HRF HbR_3_4 | 1 | max_2HRF HbT_4_3 | |
| 2 | min_2HRF HbR_6_5 | 2 | max_2HRF HbO_4_3 | |
| 3 | min_2HRF HbR_3_2 | 3 | min_3HRF HbT_3_1 | |
| 4 | std_2HRF HbT_5_3 | 4 | min_3HRF HbT_1_2 | |
| 5 | std_2HRF HbT_1_1 | 5 | min_2HRF HbR_1_1 | |
| 6 | mean_2HRF HbO_1_1 | 6 | std_3HRF HbT_3_1 | |
| 7 | std_2HRF HbT_4_3 | 7 | std_2HRF HbT_4_3 | |
| 8 | std_2HRF HbT_3_1 | 8 | max_2HRF HbR_3_2 | |
| 9 | mean_2HRF HbO_3_1 | 9 | min_3HRF HbT_1_1 | |
| 10 | max_3HRF HbT_8_6 | 10 | std_2HRF HbO_4_3 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Cervantes, A.; Herrera, V.; Zamora-Mendoza, B.N.; Flores-Ramírez, R.; López-Cano, A.A.; Guevara, E. Exploring New Horizons: fNIRS and Machine Learning in Understanding PostCOVID-19. Mach. Learn. Knowl. Extr. 2025, 7, 129. https://doi.org/10.3390/make7040129
Morales-Cervantes A, Herrera V, Zamora-Mendoza BN, Flores-Ramírez R, López-Cano AA, Guevara E. Exploring New Horizons: fNIRS and Machine Learning in Understanding PostCOVID-19. Machine Learning and Knowledge Extraction. 2025; 7(4):129. https://doi.org/10.3390/make7040129
Chicago/Turabian StyleMorales-Cervantes, Antony, Victor Herrera, Blanca Nohemí Zamora-Mendoza, Rogelio Flores-Ramírez, Aaron A. López-Cano, and Edgar Guevara. 2025. "Exploring New Horizons: fNIRS and Machine Learning in Understanding PostCOVID-19" Machine Learning and Knowledge Extraction 7, no. 4: 129. https://doi.org/10.3390/make7040129
APA StyleMorales-Cervantes, A., Herrera, V., Zamora-Mendoza, B. N., Flores-Ramírez, R., López-Cano, A. A., & Guevara, E. (2025). Exploring New Horizons: fNIRS and Machine Learning in Understanding PostCOVID-19. Machine Learning and Knowledge Extraction, 7(4), 129. https://doi.org/10.3390/make7040129