Automated Identification and Classification of Plant Species in Heterogeneous Plant Areas Using Unmanned Aerial Vehicle-Collected RGB Images and Transfer Learning
Abstract
:1. Introduction
- We utilized plant mapping in an area with a diverse range of plant species and created a dedicated image dataset for the classification of seven distinct plant types. This approach stands in contrast to relying on publicly available datasets, which may encompass a wider range but could potentially introduce irrelevant or less accurate data into our analysis. Employing UAV datasets allows us greater control over data quality, ensuring that our findings are directly relevant to the precise areas under study.
- The test results demonstrate that the fine-tuned pre-trained transfer learning model (EfficientNetV2) achieves a high classification accuracy of 99%. Such a high accuracy rate highlights the robustness and proficiency of the implemented approach in accurately identifying and distinguishing between various plant types.
- A comparative study was also conducted, comparing the EfficientNetV2 model with other widely used transfer learning models, such as the ResNet50, Xception, DenseNet121, InceptionV3, and mobileNetV2, and providing a comprehensive understanding of their strengths and weaknesses.
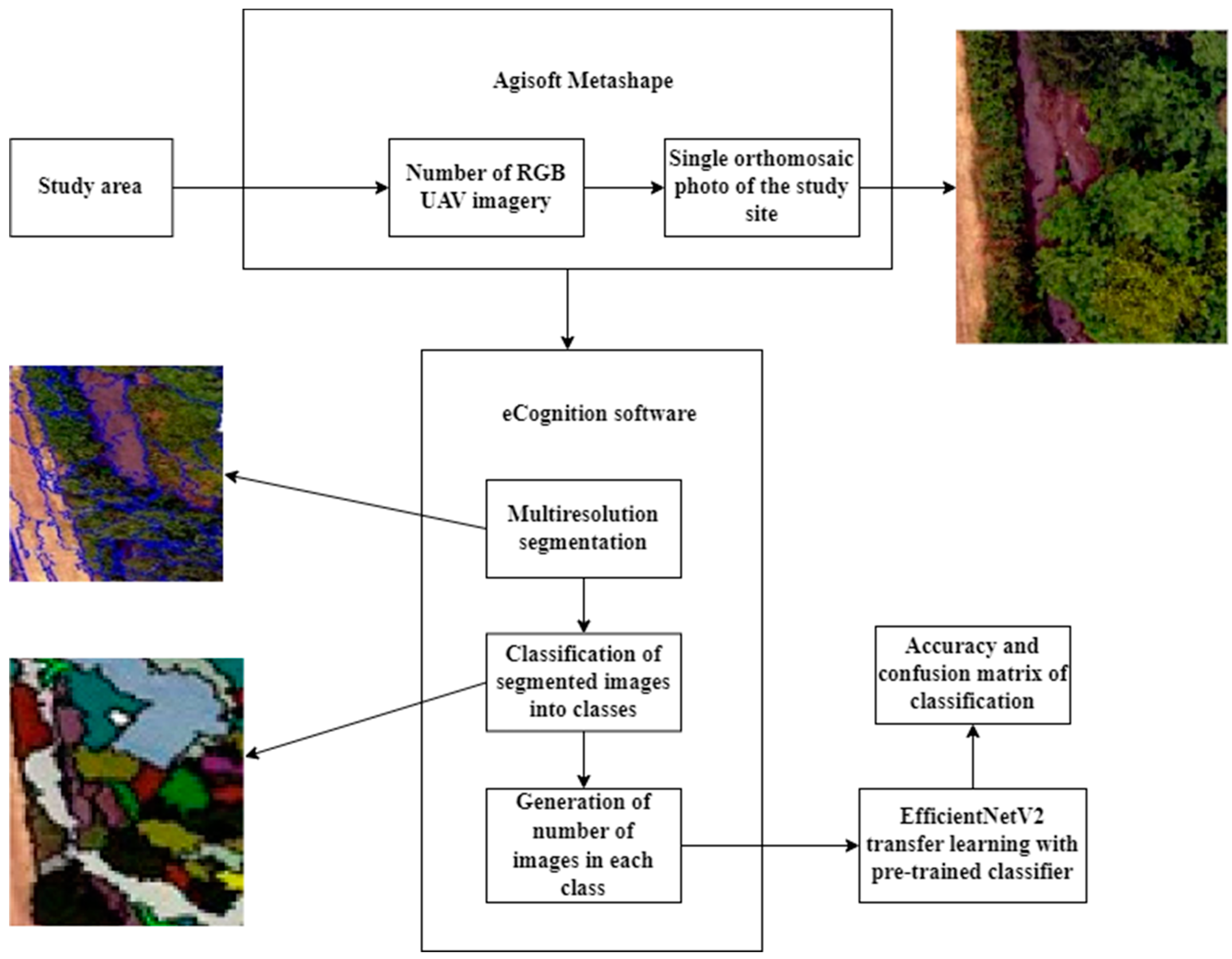
2. Materials and Methods
2.1. Study Area
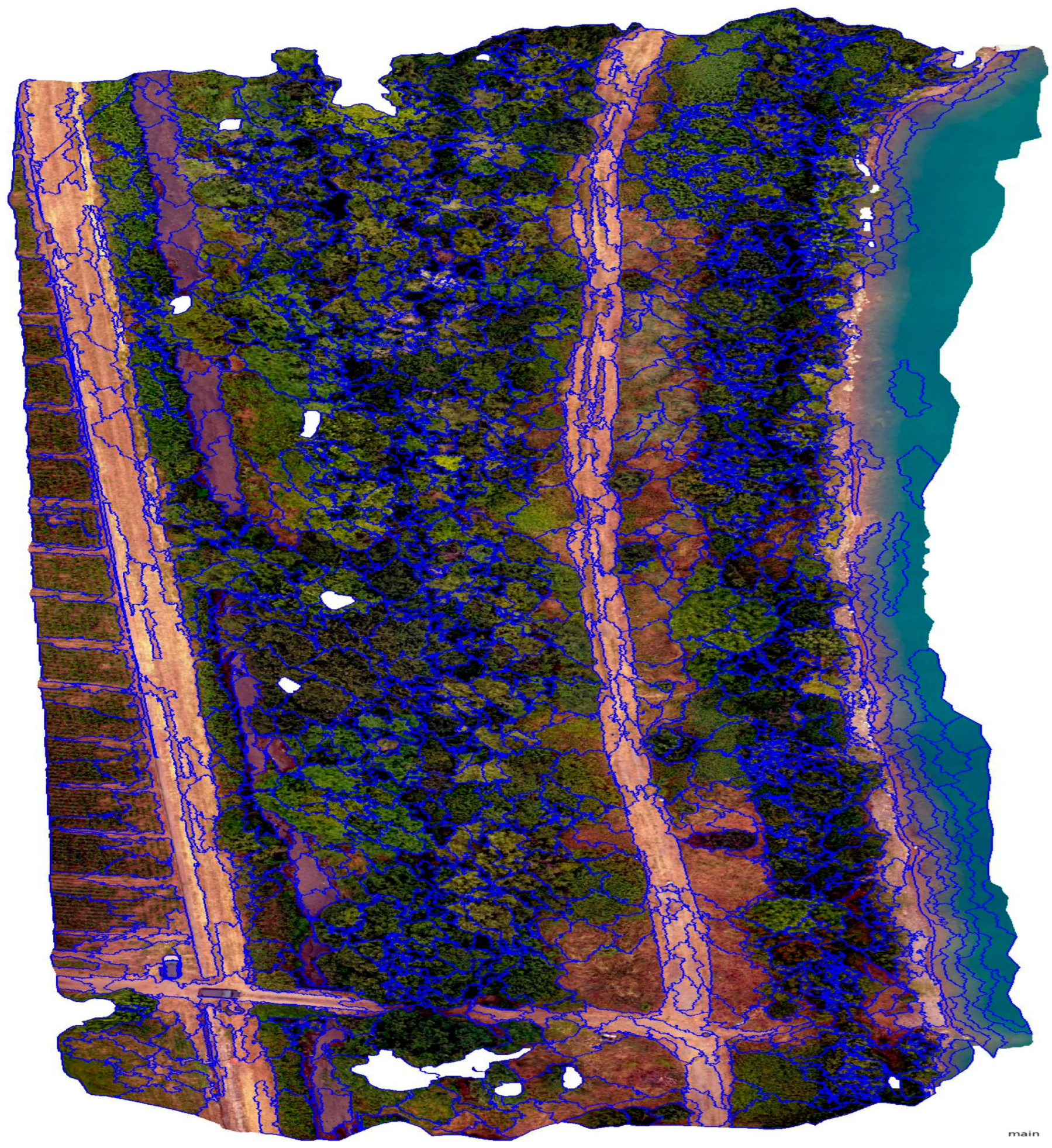
2.2. Object-Based Segmentation and Preparation of Supervised Data
- Step 1. Segmentation and Feature Considerations:
- Step 2. Training Sample Selection:
- Step 3. Supervised Learning with the KNN Algorithm:
2.3. Tree Image Extraction with Ground Truth Label
- The plant class image was imported into the ArcGIS software.
- A polygon mask was prepared to define the size of the images in the dataset and represent them with a rectangular mask. This mask was used to shift and clip the images to a larger image.
- The “Extract by Mask” tool within ArcGIS was used to clip and generate an image dataset matching the size of the polygon mask, which was tailored to each specific plant species class.
2.4. Transfer Learning: EfficientNetV2
- Take layers from a previously trained model.
- Freeze them, so as to avoid destroying any of the information they contain during future training rounds.
- Add some new, trainable layers on top of the frozen layers. They will learn to turn the old features into predictions on a new dataset.
- Train the new layers on the new dataset.
3. Results
3.1. Input Data Details
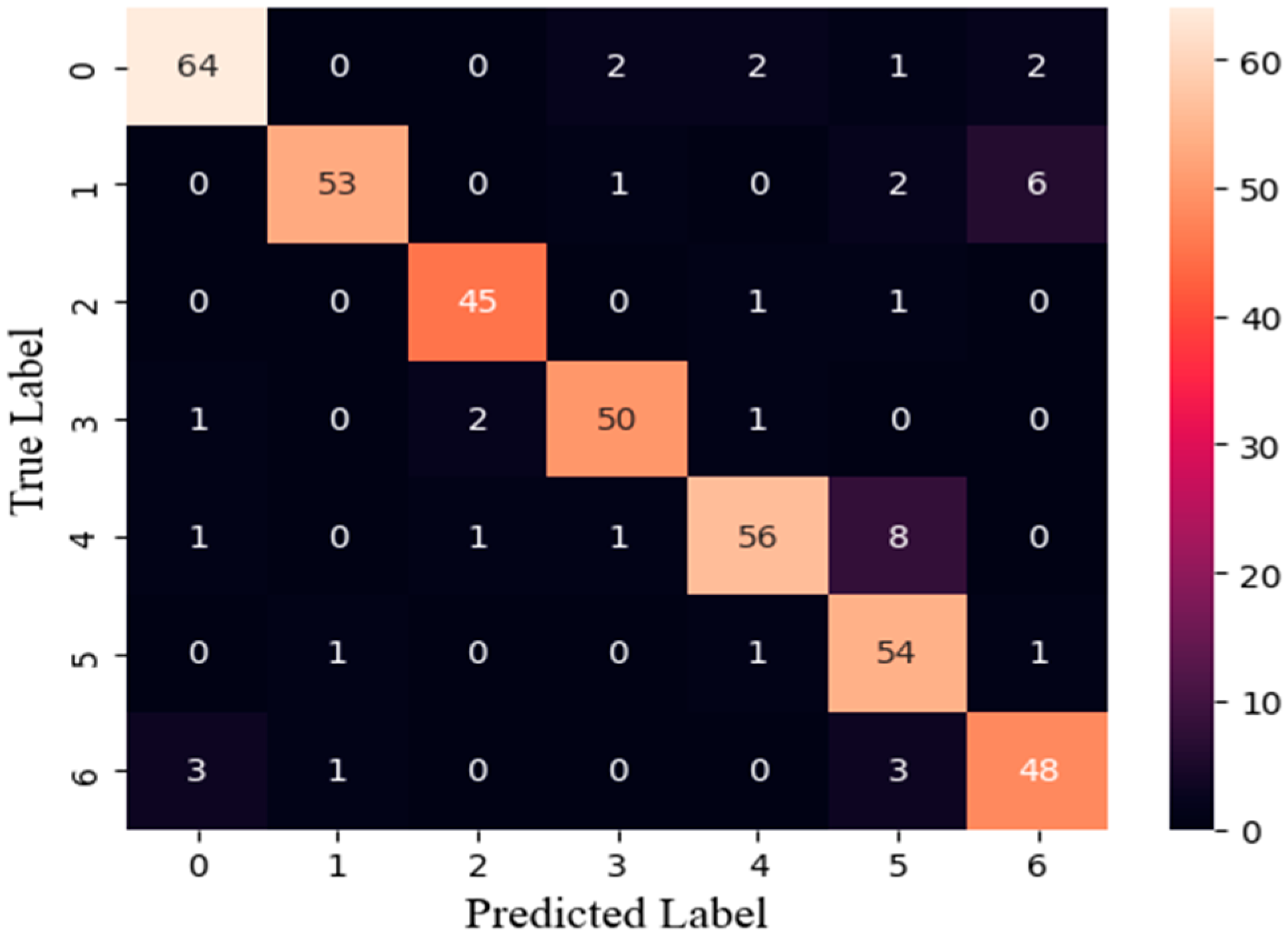
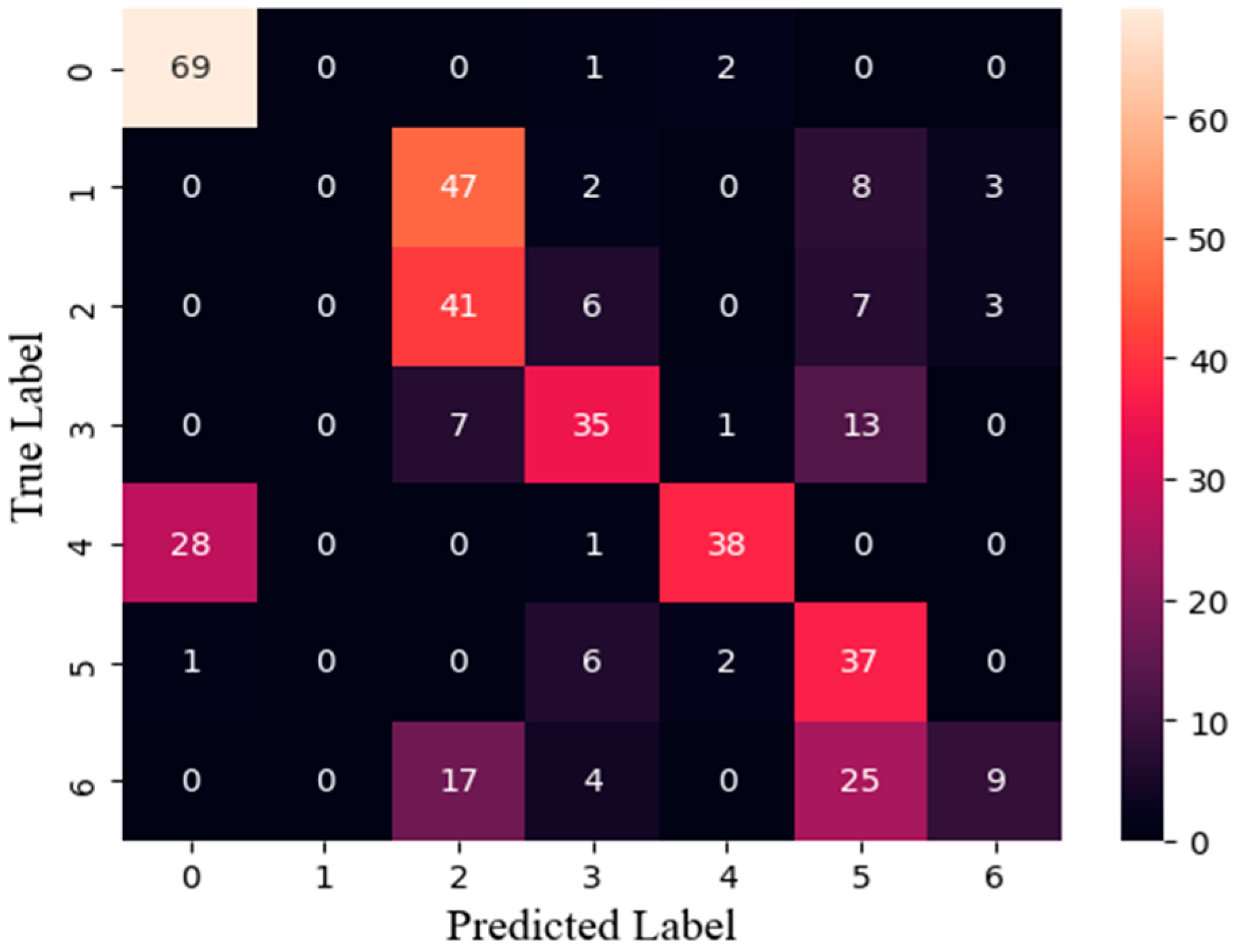
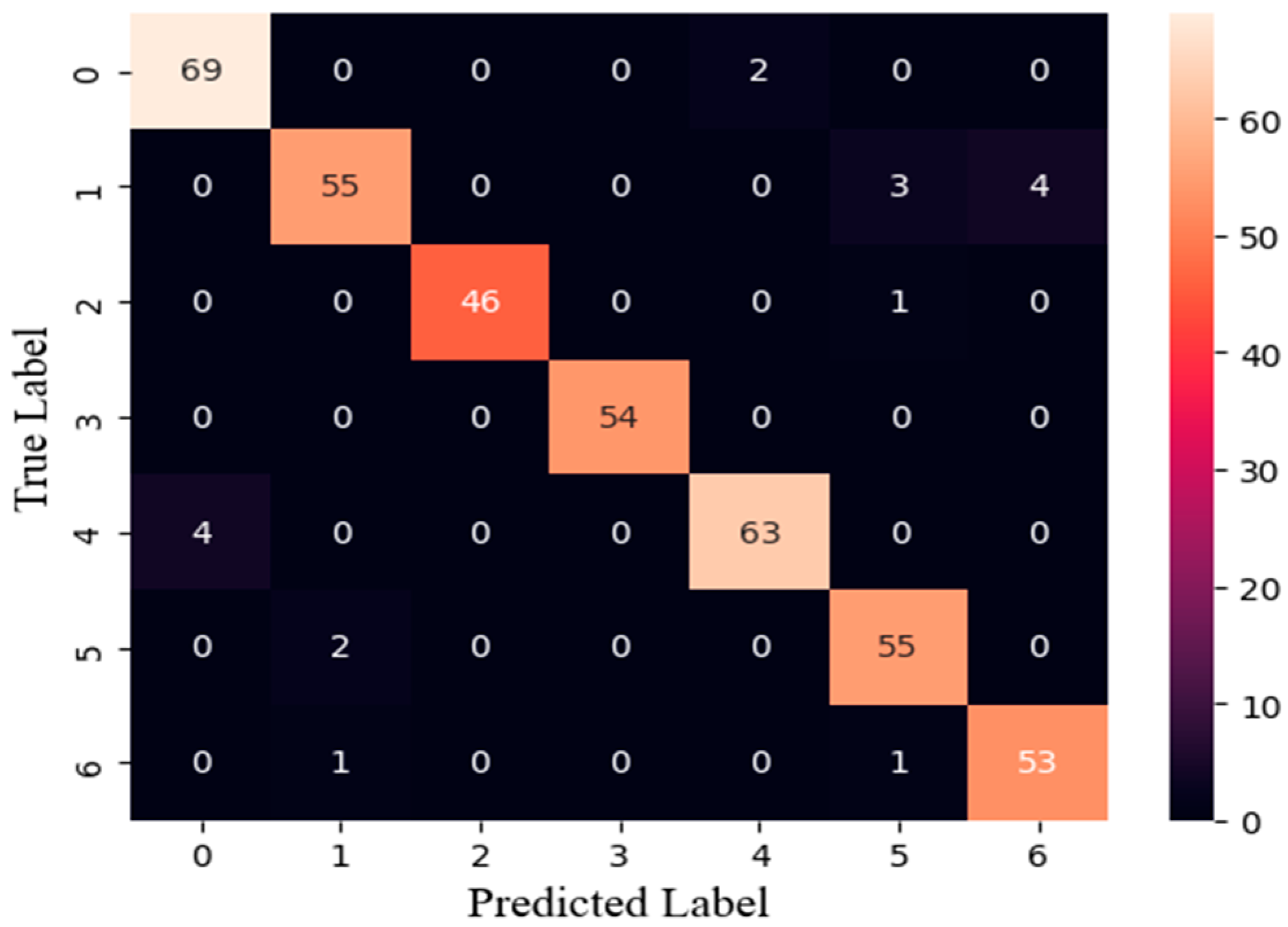
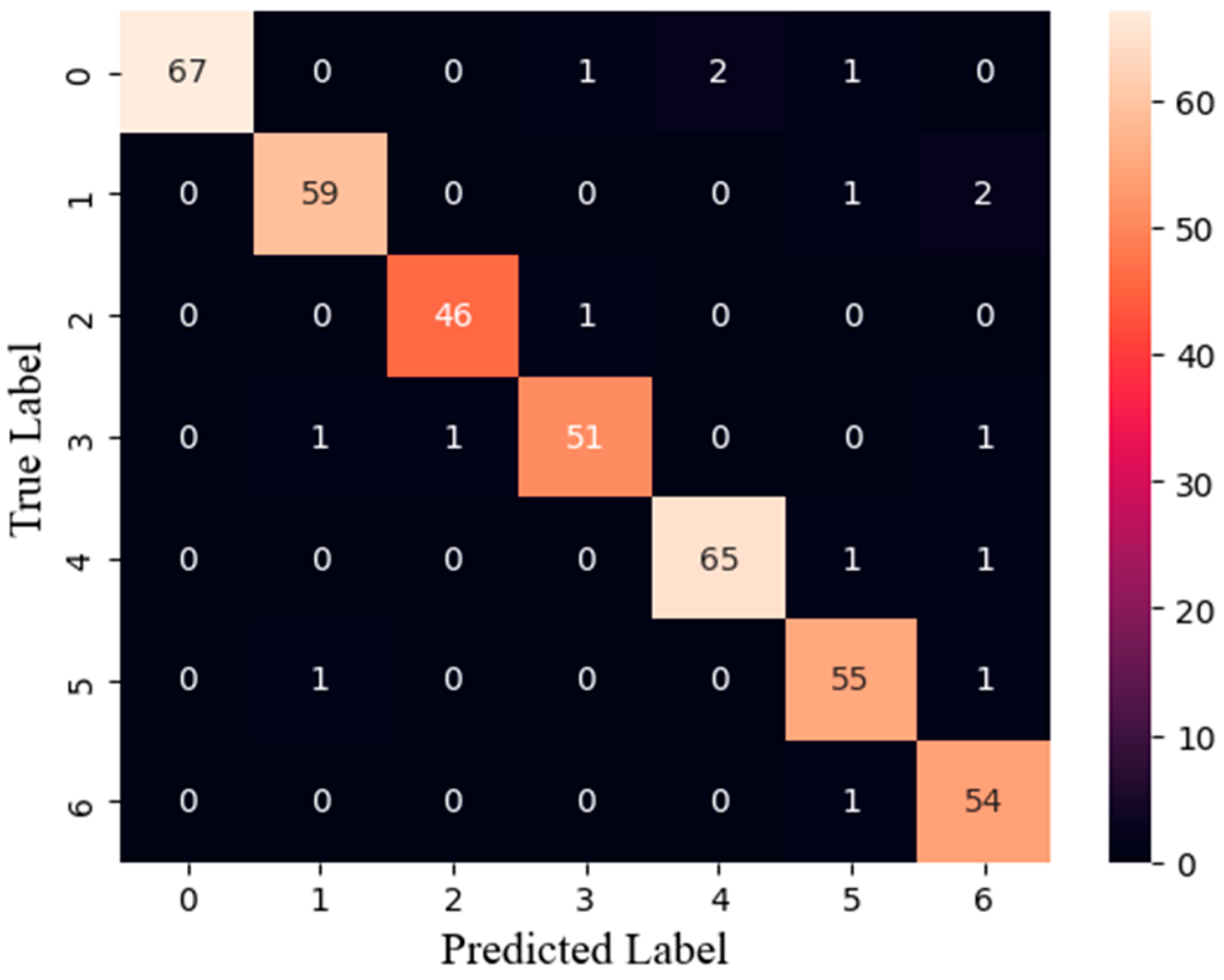
3.2. Classification Model Evaluation
3.3. Definition of the Terms
- Class 0: Standard Deviation values for channels R, G, B: [0.213517 0.2134958 0.1996962]
- Class 1: Standard Deviation values for channels R, G, B: [0.122984 0.11884958 0.08400311]
- Class 2: Standard Deviation values for channels R, G, B: [0.13345262 0.11413942 0.10716095]
- Class 3: Standard Deviation values for channels R, G, B: [0.12690945 0.13369219 0.10653751]
- Class 4: Standard Deviation values for channels R, G, B: [0.20748237 0.20688726 0.20193826]
- Class 5: Standard Deviation values for channels R, G, B: [0.15583675 0.15177113 0.12188773]
- Class 6: Standard Deviation values for channels R, G, B: [0.14056665 0.145093 0.09837905].
- Class 0: Pixel values in this class show a relatively high variability in the red and green channels compared to the blue channel.
- Class 1: Pixel values in this class show a low variability across all three color channels (R, G, B), indicating a more uniform color distribution.
- Class 2: Similarly to Class 1, pixel values in this class also show a relatively low variability across all three color channels.
- Class 3: Pixel values in this class show a moderate variability in the red and green channels, with a slightly high variability in the blue channel.
- Class 4: Pixel values in this class show a relatively high variability across all three color channels.
- Class 5: Pixel values in this class show a moderate variability in all three color channels.
- Class 6: Pixel values in this class show a moderate variability in the red and green channels, with a reduced variability in the blue channel.
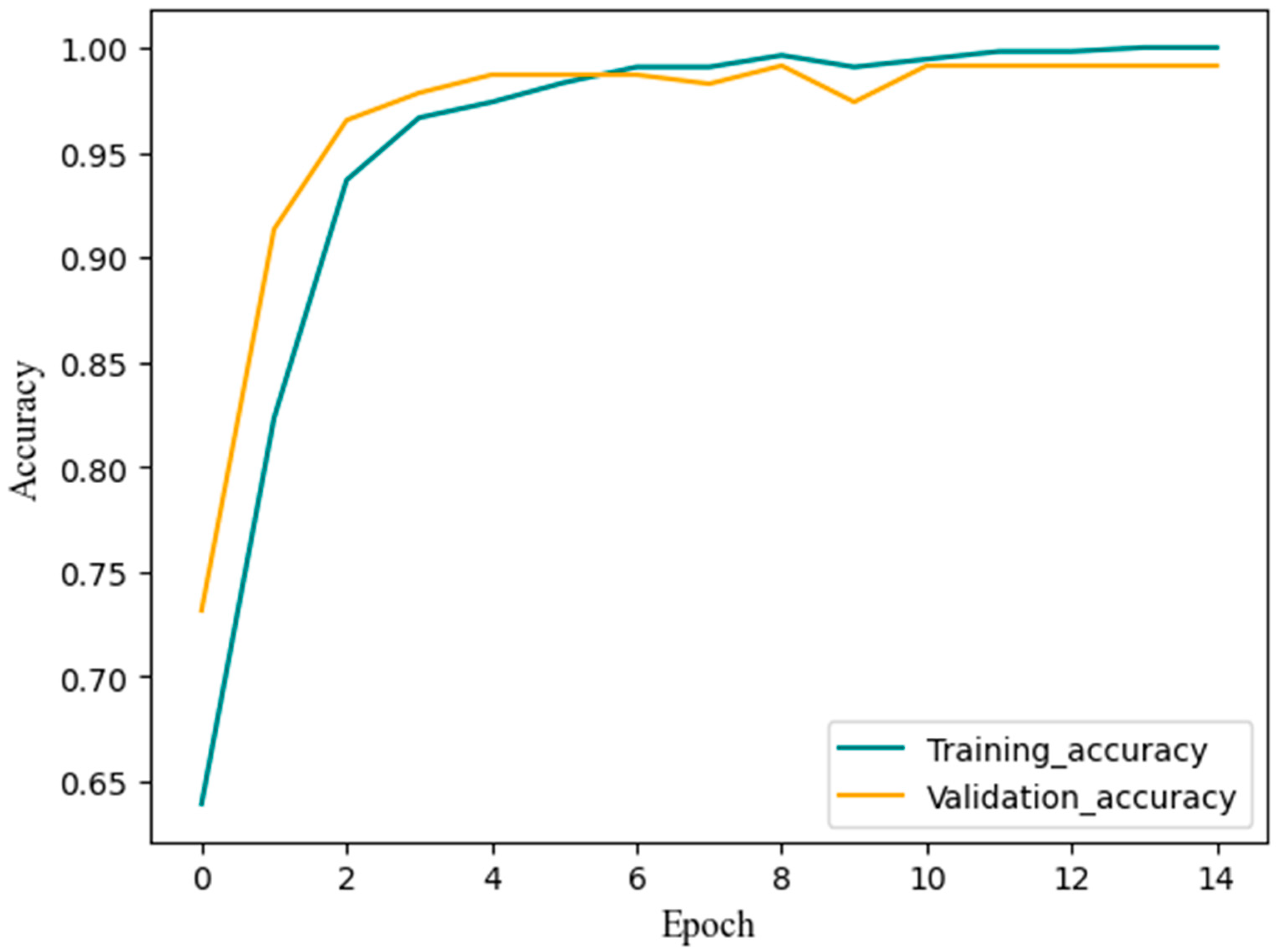
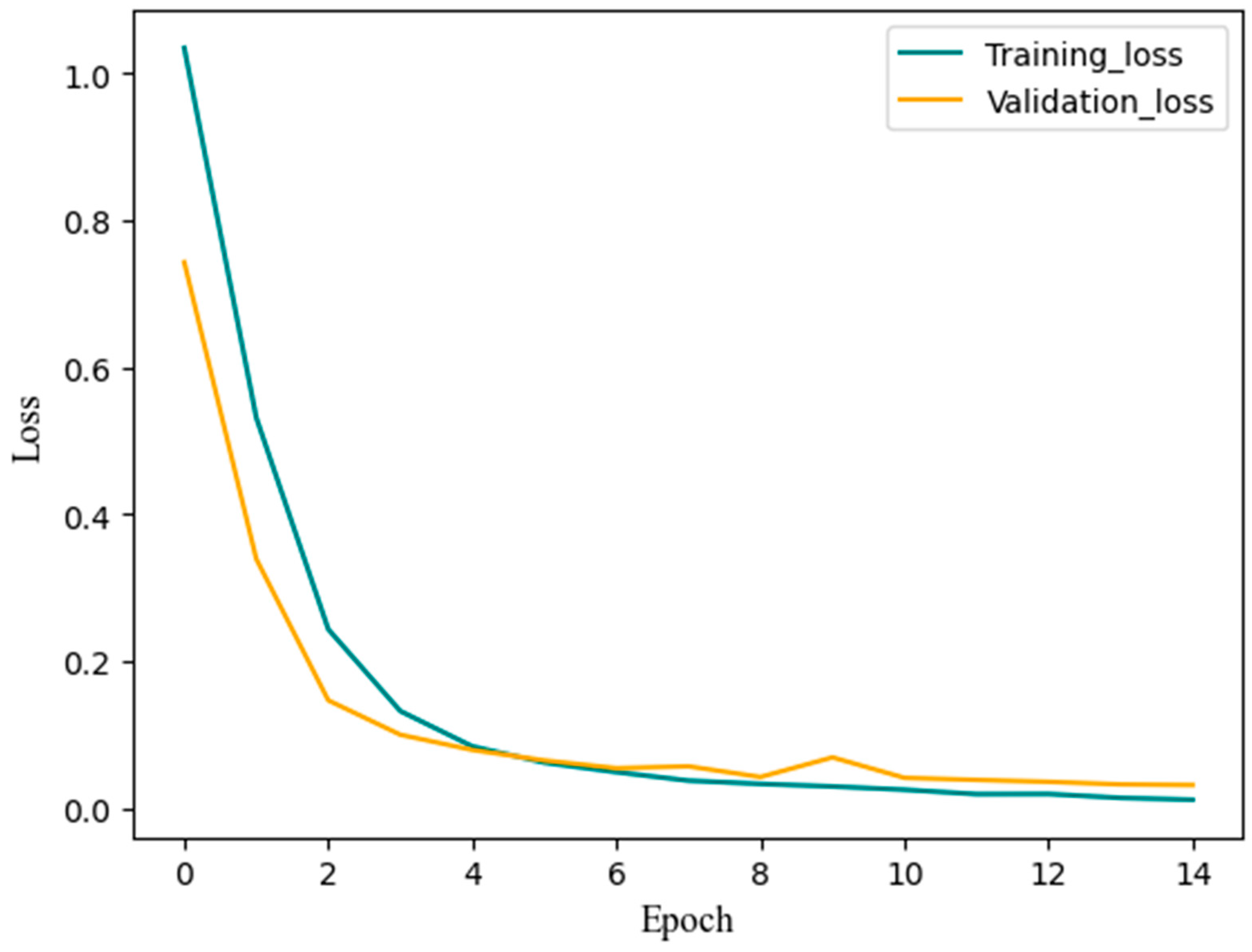
3.4. Training Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cope, J.S.; Corney, D.; Clark, J.Y.; Remagnino, P.; Wilkin, P. Plant species identification using digital morphometrics: A review. Expert Syst. Appl. 2012, 39, 7562–7573. [Google Scholar] [CrossRef]
- Grinblat, G.L.; Uzal, L.C.; Larese, M.G.; Granitto, P.M. Deep learning for plant identification using vein morphological patterns. Comput. Electron. Agric. 2016, 127, 418–424. [Google Scholar] [CrossRef]
- Dyrmann, M.; Karstoft, H.; Midtiby, H.S. Plant species classification using deep convolutional neural network. Biosyst. Eng. 2016, 151, 72–80. [Google Scholar] [CrossRef]
- Kaya, A.; Keceli, A.S.; Catal, C.; Yalic, H.Y.; Temucin, H.; Tekinerdogan, B. Analysis of transfer learning for deep neural network based plant classification models. Comput. Electron. Agric. 2019, 158, 20–29. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Comput. Electron. Agric. 2018, 153, 46–53. [Google Scholar] [CrossRef]
- Chen, F.; Tsou, J.Y. Assessing the effects of convolutional neural network architectural factors on model performance for remote sensing image classification: An in-depth investigation. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102865. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D.D. A survey of transfer learning. J. Big Data 2016, 3, 1345–1459. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.; Zhang, D.; Sun, Y.; Nanehkaran, Y. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. 2020, 173, 105393. [Google Scholar] [CrossRef]
- Ahmadibeni, A.; Jones, B.; Shirkhodaie, A. Transfer learning from simulated SAR imagery using multi-output convolutional neural networks. In Applications of Machine Learning 2020, Presented at the SPIE Optical Engineering + Applications, Online, CA, USA, 24 August–4 September 2020; Zelinski, M.E., Taha, T.M., Howe, J., Awwal, A.A., Iftekharuddin, K.M., Eds.; SPIE: Philadelphia, PA, USA, 2020; p. 30. [Google Scholar] [CrossRef]
- Fyleris, T.; Kriščiūnas, A.; Gružauskas, V.; Čalnerytė, D.; Barauskas, R. Urban Change Detection from Aerial Images Using Convolutional Neural Networks and Transfer Learning. ISPRS Int. J. Geo-Inf. 2022, 11, 246. [Google Scholar] [CrossRef]
- Liu, J.; Chen, K.; Xu, G.; Sun, X.; Yan, M.; Diao, W.; Han, H. Convolutional Neural Network-Based Transfer Learning for Optical Aerial Images Change Detection. IEEE Geosci. Remote Sens. Lett. 2020, 17, 127–131. [Google Scholar] [CrossRef]
- Rostami, M.; Kolouri, S.; Eaton, E.; Kim, K. Deep Transfer Learning for Few-Shot SAR Image Classification. Remote Sens. 2019, 11, 1374. [Google Scholar] [CrossRef]
- Bin Tufail, A.; Ullah, I.; Khan, R.; Ali, L.; Yousaf, A.; Rehman, A.U.; Alhakami, W.; Hamam, H.; Cheikhrouhou, O.; Ma, Y.-K. Recognition of Ziziphus lotus through Aerial Imaging and Deep Transfer Learning Approach. Mob. Inf. Syst. 2021, 2021, 4310321. [Google Scholar] [CrossRef]
- Lu, Y.; Young, S. A survey of public datasets for computer vision tasks in precision agriculture. Comput. Electron. Agric. 2020, 178, 105760. [Google Scholar] [CrossRef]
- Hunt, E.R., Jr.; Daughtry, C.S.T. What good are unmanned aircraft systems for agricultural remote sensing and precision agriculture? Int. J. Remote Sens. 2018, 39, 5345–5376. [Google Scholar] [CrossRef]
- Bouguettaya, A.; Zarzour, H.; Kechida, A.; Taberkit, A.M. Deep learning techniques to classify agricultural crops through UAV imagery: A review. Neural Comput. Appl. 2022, 34, 9511–9536. [Google Scholar] [CrossRef]
- Kentsch, S.; Lopez Caceres, M.L.; Serrano, D.; Roure, F.; Diez, Y. Computer Vision and Deep Learning Techniques for the Analysis of Drone-Acquired Forest Images, a Transfer Learning Study. Remote Sens. 2020, 12, 1287. [Google Scholar] [CrossRef]
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Heaphy, M.; Dungey, H.S. Assessing very high resolution UAV imagery for monitoring forest health during a simulated disease outbreak. ISPRS J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Torresan, C.; Berton, A.; Carotenuto, F.; Di Gennaro, S.F.; Gioli, B.; Matese, A.; Miglietta, F.; Vagnoli, C.; Zaldei, A.; Wallace, L. Forestry applications of UAVs in Europe: A review. Int. J. Remote Sens. 2017, 38, 2427–2447. [Google Scholar] [CrossRef]
- Xu, W.; Luo, W.; Zhang, C.; Zhao, X.; von Gadow, K.; Zhang, Z. Biodiversity-ecosystem functioning relationships of overstorey versus understorey trees in an old-growth temperate forest. Ann. For. Sci. 2019, 76, 64. [Google Scholar] [CrossRef]
- Nasiri, V.; Darvishsefat, A.A.; Arefi, H.; Griess, V.C.; Sadeghi, S.M.M.; Borz, S.A. Modeling Forest Canopy Cover: A Synergistic Use of Sentinel-2, Aerial Photogrammetry Data, and Machine Learning. Remote Sens. 2022, 14, 1453. [Google Scholar] [CrossRef]
- Onishi, M.; Ise, T. Explainable identification and mapping of trees using UAV RGB image and deep learning. Sci. Rep. 2021, 11, 903. [Google Scholar] [CrossRef] [PubMed]
- Akcay, O.; Avsar, E.O.; Inalpulat, M.; Genc, L.; Cam, A. Assessment of Segmentation Parameters for Object-Based Land Cover Classification Using Color-Infrared Imagery. ISPRS Int. J. Geo-Inf. 2018, 7, 424. [Google Scholar] [CrossRef]
- Gašparović, M.; Zrinjski, M.; Barković, Đ.; Radočaj, D. An automatic method for weed mapping in oat fields based on UAV imagery. Comput. Electron. Agric. 2020, 173, 105385. [Google Scholar] [CrossRef]
- Pandey, A.; Jain, K. An intelligent system for crop identification and classification from UAV images using conjugated dense convolutional neural network. Comput. Electron. Agric. 2022, 192, 106543. [Google Scholar] [CrossRef]
- Reedha, R.; Dericquebourg, E.; Canals, R.; Hafiane, A. Transformer Neural Network for Weed and Crop Classification of High Resolution UAV Images. Remote Sens. 2022, 14, 592. [Google Scholar] [CrossRef]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef]
- Benz, U.C.; Hofmann, P.; Willhauck, G.; Lingenfelder, I.; Heynen, M. Multi-resolution, object-oriented fuzzy analysis of remote sensing data for GIS-ready information. ISPRS J. Photogramm. Remote Sens. 2004, 58, 239–258. [Google Scholar] [CrossRef]
- Ma, L.; Li, M.; Ma, X.; Cheng, L.; Du, P.; Liu, Y. A review of supervised object-based land-cover image classification. ISPRS J. Photogramm. Remote Sens. 2017, 130, 277–293. [Google Scholar] [CrossRef]
- Deng, Z.; Zhu, X.; Cheng, D.; Zong, M.; Zhang, S. Efficient kNN classification algorithm for big data. Neurocomputing 2016, 195, 143–148. [Google Scholar] [CrossRef]
- Mehmood, T.; Gerevini, A.; Lavelli, A.; Serina, I. Leveraging Multi-task Learning for Biomedical Named Entity Recognition. In AI*IA 2019—Advances in Artificial Intelligence; Alviano, M., Greco, G., Scarcello, F., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; pp. 431–444. [Google Scholar]
- Mehmood, T.; Gerevini, A.E.; Lavelli, A.; Serina, I. Combining Multi-task Learning with Transfer Learning for Biomedical Named Entity Recognition. Procedia Comput. Sci. 2020, 176, 848–857. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNetV2: Smaller Models and Faster Training. arXiv 2021, arXiv:2104.00298. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27 June–28 July 2016; pp. 2818–2826. Available online: https://www.cv-foundation.org/openaccess/content_cvpr_2016/html/Szegedy_Rethinking_the_Inception_CVPR_2016_paper.html (accessed on 22 April 2023).
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2019, arXiv:1801.04381. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. arXiv 2017, arXiv:1610.02357. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:1608.06993. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27 June–28 July 2016; pp. 770–778. Available online: https://openaccess.thecvf.com/content_cvpr_2016/html/He_Deep_Residual_Learning_CVPR_2016_paper.html (accessed on 22 April 2023).
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2020, arXiv:1905.11946. [Google Scholar] [CrossRef]






| Species | Total No. of Images | No. of Test Images | No. of Training Images |
|---|---|---|---|
| Agropyron repens | 215 | 71 | 144 |
| Ailanthus altissima | 193 | 62 | 131 |
| Arrhenatherum elatius | 174 | 47 | 127 |
| Artemisia verlotiorum | 194 | 54 | 140 |
| Populus nigra | 208 | 67 | 141 |
| Rubus caesius | 182 | 57 | 125 |
| Ulmus minor | 208 | 55 | 153 |
| Total | 1374 | 413 | 961 |
| Species | Precision | Recall | Accuracy | F1-Score |
|---|---|---|---|---|
| Agropyron repens | 1.0 | 0.97 | 0.97 | 0.99 |
| Ailanthus altissima | 1.0 | 1.0 | 1.0 | 1.0 |
| Arrhenatherum elatius | 0.98 | 1.0 | 1.0 | 0.99 |
| Artemisia verlotiorum | 1.0 | 0.98 | 0.98 | 0.99 |
| Populus nigra | 0.97 | 1.0 | 1.0 | 0.99 |
| Rubus caesius | 1.0 | 1.0 | 1.0 | 1.0 |
| Ulmus minor | 1.0 | 1.0 | 1.0 | 1.0 |
| Pre-trained Model | Pre-trained Dataset | Accuracy (%) | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| EfficientNetV2 | ImageNet | 99.2 | 0.992 | 0.993 | 0.993 |
| MobileNetV2 | ImageNet | 96.6 | 0.967 | 0.964 | 0.965 |
| Xception | ImageNet | 0.956 | 0.955 | 0.958 | 0.954 |
| DensNet121 | ImageNet | 0.953 | 0.951 | 0.953 | 0.951 |
| InceptionV3 | ImageNet | 91.5 | 0.916 | 0.918 | 0.915 |
| ResNet50 | ImageNet | 60.2 | 0.655 | 0.615 | 0.594 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tariku, G.; Ghiglieno, I.; Gilioli, G.; Gentilin, F.; Armiraglio, S.; Serina, I. Automated Identification and Classification of Plant Species in Heterogeneous Plant Areas Using Unmanned Aerial Vehicle-Collected RGB Images and Transfer Learning. Drones 2023, 7, 599. https://doi.org/10.3390/drones7100599
Tariku G, Ghiglieno I, Gilioli G, Gentilin F, Armiraglio S, Serina I. Automated Identification and Classification of Plant Species in Heterogeneous Plant Areas Using Unmanned Aerial Vehicle-Collected RGB Images and Transfer Learning. Drones. 2023; 7(10):599. https://doi.org/10.3390/drones7100599
Chicago/Turabian StyleTariku, Girma, Isabella Ghiglieno, Gianni Gilioli, Fulvio Gentilin, Stefano Armiraglio, and Ivan Serina. 2023. "Automated Identification and Classification of Plant Species in Heterogeneous Plant Areas Using Unmanned Aerial Vehicle-Collected RGB Images and Transfer Learning" Drones 7, no. 10: 599. https://doi.org/10.3390/drones7100599
APA StyleTariku, G., Ghiglieno, I., Gilioli, G., Gentilin, F., Armiraglio, S., & Serina, I. (2023). Automated Identification and Classification of Plant Species in Heterogeneous Plant Areas Using Unmanned Aerial Vehicle-Collected RGB Images and Transfer Learning. Drones, 7(10), 599. https://doi.org/10.3390/drones7100599








