Molecular Communication Approaches for Wetware Artificial Life: A Workshop Report
Abstract
1. ALIFE 2023
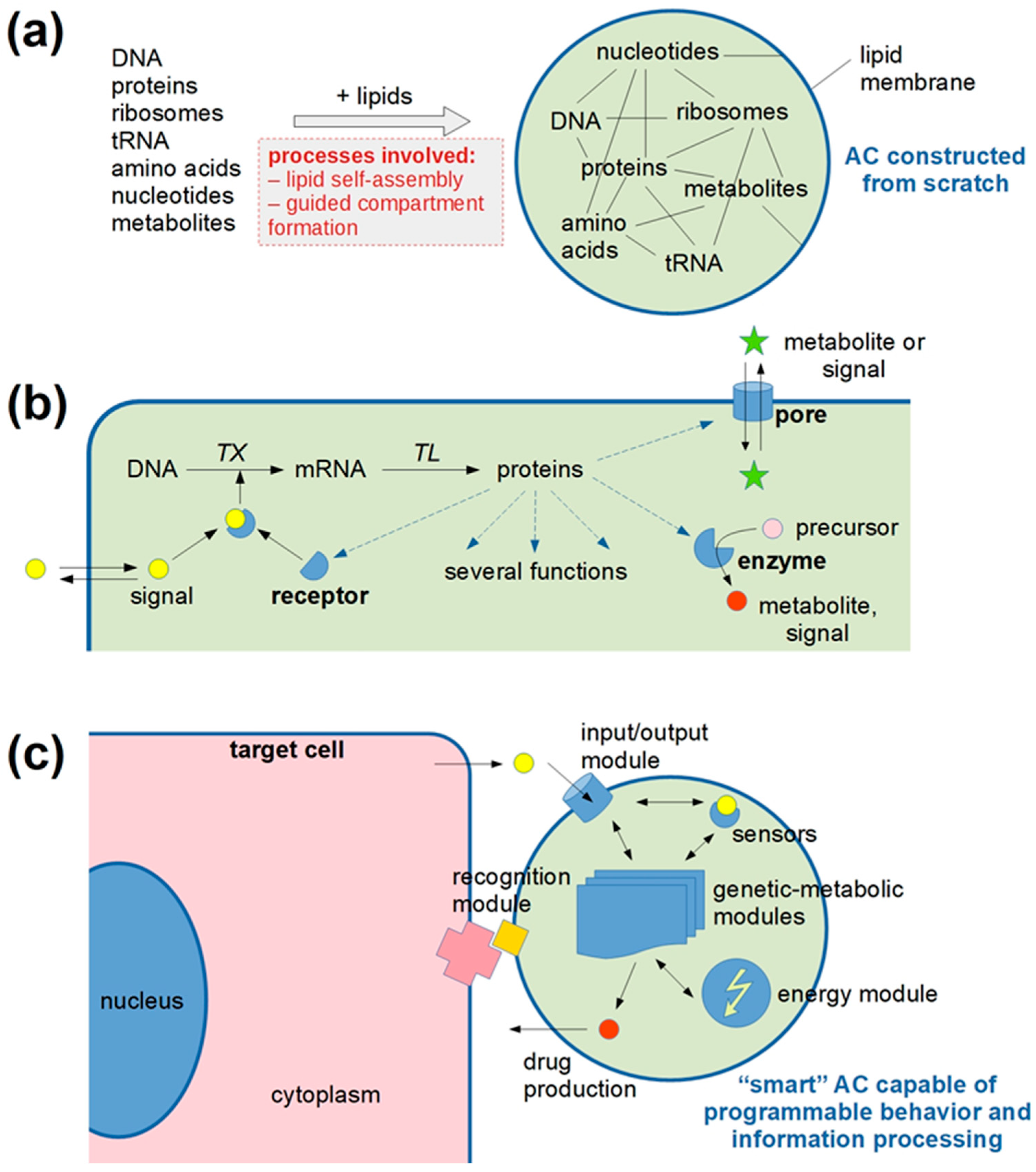
2. Synopsis of the Talks Presented at the Workshop
3. Concluding Remarks and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Molecular Communication Approaches for Wetware Artificial Life: Call for Papers
References
- Langton, C.G. Artificial Life. In Artificial Life; Langton, C.G., Ed.; Addison-Wesley: Reading, MA, USA, 1989; pp. 1–47. [Google Scholar]
- Luisi, P.L. Toward the Engineering of Minimal Living Cells. Anat. Rec. 2002, 268, 208–214. [Google Scholar] [CrossRef]
- Mansy, S.S.; Szostak, J.W. Reconstructing the Emergence of Cellular Life through the Synthesis of Model Protocells. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 47–54. [Google Scholar] [CrossRef]
- Martos, A.; Jiménez, M.; Rivas, G.; Schwille, P. Towards a Bottom-up Reconstitution of Bacterial Cell Division. Trends Cell Biol. 2012, 22, 634–643. [Google Scholar] [CrossRef]
- Gaut, N.J.; Adamala, K.P. Reconstituting Natural Cell Elements in Synthetic Cells. Adv. Biol. 2021, 5, e2000188. [Google Scholar] [CrossRef] [PubMed]
- Göpfrich, K.; Platzman, I.; Spatz, J.P. Mastering Complexity: Towards Bottom-up Construction of Multifunctional Eukaryotic Synthetic Cells. Trends Biotechnol. 2018, 36, 938–951. [Google Scholar] [CrossRef] [PubMed]
- Abil, Z.; Danelon, C. Roadmap to Building a Cell: An Evolutionary Approach. Front. Bioeng. Biotechnol. 2020, 8, 927. [Google Scholar] [CrossRef] [PubMed]
- Salehi-Reyhani, A.; Ces, O.; Elani, Y. Artificial Cell Mimics as Simplified Models for the Study of Cell Biology. Exp. Biol. Med. 2017, 242, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Buddingh’, B.C.; van Hest, J.C.M. Artificial Cells: Synthetic Compartments with Life-like Functionality and Adaptivity. Acc. Chem. Res. 2017, 50, 769–777. [Google Scholar] [CrossRef]
- Tang, T.-Y.D.; van Swaay, D.; deMello, A.; Anderson, J.L.R.; Mann, S. In Vitro Gene Expression within Membrane-Free Coacervate Protocells. Chem. Commun. 2015, 51, 11429–11432. [Google Scholar] [CrossRef]
- Kurihara, K.; Okura, Y.; Matsuo, M.; Toyota, T.; Suzuki, K.; Sugawara, T. A Recursive Vesicle-Based Model Protocell with a Primitive Model Cell Cycle. Nat. Commun. 2015, 6, 8352. [Google Scholar] [CrossRef]
- Ichihashi, N.; Matsuura, T.; Kita, H.; Sunami, T.; Suzuki, H.; Yomo, T. Constructing Partial Models of Cells. Cold Spring Harb. Perspect. Biol. 2010, 2, a004945. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, K.; Nomura, S.M.; Tsumoto, K.; Takiguchi, K. Construction of an In Vitro Model of a Living Cellular System. In The Minimal Cell: The Biophysics of Cell Compartment and the Origin of Cell Functionality; Luisi, P.L., Stano, P., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 173–193. ISBN 978-90-481-9944-0. [Google Scholar]
- Stano, P. Gene Expression Inside Liposomes: From Early Studies to Current Protocols. Chemistry 2019, 25, 7798–7814. [Google Scholar] [CrossRef] [PubMed]
- Stano, P. Minimal Cells: Relevance and Interplay of Physical and Biochemical Factors. Biotechnol. J. 2011, 6, 850–859. [Google Scholar] [CrossRef]
- Sato, W.; Zajkowski, T.; Moser, F.; Adamala, K.P. Synthetic Cells in Biomedical Applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2022, 14, e1761. [Google Scholar] [CrossRef]
- Stano, P.; Gentili, P.L.; Damiano, L.; Magarini, M. A Role for Bottom-Up Synthetic Cells in the Internet of Bio-Nano Things? Molecules 2023, 28, 5564. [Google Scholar] [CrossRef]
- Krinsky, N.; Kaduri, M.; Zinger, A.; Shainsky-Roitman, J.; Goldfeder, M.; Benhar, I.; Hershkovitz, D.; Schroeder, A. Synthetic Cells Synthesize Therapeutic Proteins inside Tumors. Adv. Healthc. Mater. 2018, 7, e1701163. [Google Scholar] [CrossRef]
- Westensee, I.N.; Städler, B. Artificial Cells Eavesdropping on HepG2 Cells. Interface Focus 2023, 13, 20230007. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, D.; Chowdhry, R.; Smith, J.M.; Booth, M.J. Orthogonal Light-Activated DNA for Patterned Biocomputing within Synthetic Cells. J. Am. Chem. Soc. 2023, 145, 9471–9480. [Google Scholar] [CrossRef]
- Chen, G.; Levin, R.; Landau, S.; Kaduri, M.; Adir, O.; Ianovici, I.; Krinsky, N.; Doppelt-Flikshtain, O.; Shklover, J.; Shainsky-Roitman, J.; et al. Implanted Synthetic Cells Trigger Tissue Angiogenesis through de Novo Production of Recombinant Growth Factors. Proc. Natl. Acad. Sci. USA 2022, 119, e2207525119. [Google Scholar] [CrossRef]
- Wu, F.; Tan, C. The Engineering of Artificial Cellular Nanosystems Using Synthetic Biology Approaches. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014, 6, 369–383. [Google Scholar] [CrossRef]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.-Y.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science 2010, 329, 52–56. [Google Scholar] [CrossRef]
- Glass, J.I. Synthetic Genomics and the Construction of a Synthetic Bacterial Cell. Perspect. Biol. Med. 2012, 55, 473–489. [Google Scholar] [CrossRef]
- Hutchison, C.A.; Chuang, R.-Y.; Noskov, V.N.; Assad-Garcia, N.; Deerinck, T.J.; Ellisman, M.H.; Gill, J.; Kannan, K.; Karas, B.J.; Ma, L.; et al. Design and Synthesis of a Minimal Bacterial Genome. Science 2016, 351, aad6253. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.F.; Sun, L.; Wise, K.S.; Assad-Garcia, N.; Karas, B.J.; Deerinck, T.J.; Ellisman, M.H.; Mershin, A.; Gershenfeld, N.; Chuang, R.-Y.; et al. Genetic Requirements for Cell Division in a Genomically Minimal Cell. Cell 2021, 184, 2430–2440.e16. [Google Scholar] [CrossRef] [PubMed]
- van de Cauter, L.; van Buren, L.; Koenderink, G.H.; Ganzinger, K.A. Exploring Giant Unilamellar Vesicle Production for Artificial Cells—Current Challenges and Future Directions. Small Methods 2023, 7, e2300416. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Sakuma, Y.; Kurisu, M.; Walde, P. From Vesicles toward Protocells and Minimal Cells. Soft Matter 2022, 18, 4823–4849. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Mann, S. Membranized Coacervate Microdroplets: From Versatile Protocell Models to Cytomimetic Materials. Acc. Chem. Res. 2023, 56, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Stano, P. Commentary: Rapid and Facile Preparation of Giant Vesicles by the Droplet Transfer Method for Artificial Cell Construction. Front. Bioeng. Biotechnol. 2022, 10, 1037809. [Google Scholar] [CrossRef]
- Tan, S.; Ai, Y.; Yin, X.; Xue, Z.; Fang, X.; Liang, Q.; Gong, X.; Dai, X. Recent Advances in Microfluidic Technologies for the Construction of Artificial Cells. Adv. Funct. Mater. 2023, 33, 2305071. [Google Scholar] [CrossRef]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-Free Translation Reconstituted with Purified Components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef]
- Kuruma, Y.; Ueda, T. The PURE System for the Cell-Free Synthesis of Membrane Proteins. Nat. Protoc. 2015, 10, 1328–1344. [Google Scholar] [CrossRef] [PubMed]
- Hiyama, S.; Moritani, Y.; Suda, T.; Egashira, R.; Enamoto, A.; Moore, M.; Nakano, T. Molecular Communications. In Technical Proceedings of the 2005 NSTI Nanotechnology Conference and Trade Show (Anaheim, 8–12 May 2005); TechConnect Briefs: Summerville, SC, USA, 2005; Volume 3, pp. 391–394. ISBN 0-9767985-2-2. [Google Scholar]
- Nakano, T.; Moore, M.; Enomoto, A.; Suda, T. Molecular Communication Technology as a Biological ICT. In Biological Functions for Information and Communication Technologies; Sawai, H., Ed.; Studies in Computational Intelligence; Springer: Berlin/Heidelberg, Germany, 2011; pp. 49–86. ISBN 978-3-642-15101-9. [Google Scholar]
- Nakano, T. Molecular Communication: A 10 Year Retrospective. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2017, 3, 71–78. [Google Scholar] [CrossRef]
- Akyildiz, I.F.; Pierobon, M.; Balasubramaniam, S.; Koucheryavy, Y. The Internet of Bio-Nano Things. IEEE Commun. Mag. 2015, 53, 32–40. [Google Scholar] [CrossRef]
- Kuscu, M.; Unluturk, B.D. Internet of Bio-Nano Things: A Review of Applications, Enabling Technologies and Key Challenges. ITU J. Future Evol. Technol. 2021, 2, 1–24. [Google Scholar] [CrossRef]
- Hoffmann, P.M. Life’s Ratchet. How Molecular Machines Extract Order from Chaos, 1st ed.; Basic Books: A Member of the Perseus Books Group: New York, NY, USA, 2012; ISBN 978-0-465-02253-3. [Google Scholar]
- Phillips, R.; Quake, S.R. The Biological Frontier of Physics. Phys. Today 2006, 59, 38–43. [Google Scholar] [CrossRef]
- Maturana, H.R.; Varela, F.J. Autopoiesis and Cognition: The Realization of the Living; D. Reidel Publishing Company: Dordrecht, The Netherlands, 1980; ISBN 90-277-1016-3. [Google Scholar]
- Luisi, P.L. Autopoiesis: A Review and a Reappraisal. Naturwissenschaften 2003, 90, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Stano, P.; Nehaniv, C.; Ikegami, T.; Damiano, L.; Witkowski, O. Autopoiesis: Foundations of Life, Cognition, and Emergence of Self/Other. BioSystems 2023, 232, 105008. [Google Scholar] [CrossRef] [PubMed]
- Damiano, L.; Stano, P. Synthetic Biology and Artificial Intelligence. Grounding a Cross-Disciplinary Approach to the Synthetic Exploration of (Embodied) Cognition. Complex Syst. 2018, 27, 199–228. [Google Scholar] [CrossRef]
- Damiano, L.; Stano, P. A Wetware Embodied AI? Towards an Autopoietic Organizational Approach Grounded in Synthetic Biology. Front. Bioeng. Biotechnol. 2021, 9, 873. [Google Scholar] [CrossRef]
- Damiano, L.; Stano, P. Explorative Synthetic Biology in AI. Criteria of Relevance and a Taxonomy for Synthetic Models of Living and Cognitive Processes. Artif. Life 2023, 29, 367–387. [Google Scholar] [CrossRef]
- Stano, P.; Rampioni, G.; Carrara, P.; Damiano, L.; Leoni, L.; Luisi, P.L. Semi-Synthetic Minimal Cells as a Tool for Biochemical ICT. BioSystems 2012, 109, 24–34. [Google Scholar] [CrossRef]
- Varela, F.J. Principles of Biological Autonomy; Elsevier North-Holland, Inc.: New York, NY, USA, 1979. [Google Scholar]
- LeDuc, P.R.; Wong, M.S.; Ferreira, P.M.; Groff, R.E.; Haslinger, K.; Koonce, M.P.; Lee, W.Y.; Love, J.C.; McCammon, J.A.; Monteiro-Riviere, N.A.; et al. Towards an in Vivo Biologically Inspired Nanofactory. Nat. Nanotechnol. 2007, 2, 3–7. [Google Scholar] [CrossRef]
- Cronin, L.; Krasnogor, N.; Davis, B.G.; Alexander, C.; Robertson, N.; Steinke, J.H.G.; Schroeder, S.L.M.; Khlobystov, A.N.; Cooper, G.; Gardner, P.M.; et al. The Imitation Game—A Computational Chemical Approach to Recognizing Life. Nat. Biotechnol. 2006, 24, 1203–1206. [Google Scholar] [CrossRef]
- Gardner, P.M.; Winzer, K.; Davis, B.G. Sugar Synthesis in a Protocellular Model Leads to a Cell Signalling Response in Bacteria. Nature Chem. 2009, 1, 377–383. [Google Scholar] [CrossRef]
- Lentini, R.; Santero, S.P.; Chizzolini, F.; Cecchi, D.; Fontana, J.; Marchioretto, M.; Del Bianco, C.; Terrell, J.L.; Spencer, A.C.; Martini, L.; et al. Integrating Artificial with Natural Cells to Translate Chemical Messages That Direct E. coli Behaviour. Nat. Commun. 2014, 5, 4012. [Google Scholar] [CrossRef]
- Adamala, K.P.; Martin-Alarcon, D.A.; Guthrie-Honea, K.R.; Boyden, E.S. Engineering Genetic Circuit Interactions within and between Synthetic Minimal Cells. Nat. Chem. 2017, 9, 431–439. [Google Scholar] [CrossRef]
- Rampioni, G.; D’Angelo, F.; Messina, M.; Zennaro, A.; Kuruma, Y.; Tofani, D.; Leoni, L.; Stano, P. Synthetic Cells Produce a Quorum Sensing Chemical Signal Perceived by Pseudomonas aeruginosa. Chem. Commun. 2018, 54, 2090–2093. [Google Scholar] [CrossRef]
- Ding, Y.; Contreras-Llano, L.E.; Morris, E.; Mao, M.; Tan, C. Minimizing Context Dependency of Gene Networks Using Artificial Cells. ACS Appl. Mater. Interfaces 2018, 10, 30137–30146. [Google Scholar] [CrossRef] [PubMed]
- Aufinger, L.; Simmel, F.C. Establishing Communication Between Artificial Cells. Chem.—A Eur. J. 2019, 25, 12659–12670. [Google Scholar] [CrossRef] [PubMed]
- Dubuc, E.; Pieters, P.A.; van der Linden, A.J.; van Hest, J.C.; Huck, W.T.; de Greef, T.F. Cell-Free Microcompartmentalised Transcription–Translation for the Prototyping of Synthetic Communication Networks. Curr. Opin. Biotechnol. 2019, 58, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tian, L.; Ren, Y.; Zhao, Z.; Du, H.; Zhang, Z.; Drinkwater, B.W.; Mann, S.; Han, X. Chemical Information Exchange in Organized Protocells and Natural Cell Assemblies with Controllable Spatial Positions. Small 2020, 16, e1906394. [Google Scholar] [CrossRef]
- Buddingh’, B.C.; Elzinga, J.; van Hest, J.C.M. Intercellular Communication between Artificial Cells by Allosteric Amplification of a Molecular Signal. Nat. Commun. 2020, 11, 1652. [Google Scholar] [CrossRef]
- Mukwaya, V.; Mann, S.; Dou, H. Chemical Communication at the Synthetic Cell/Living Cell Interface. Commun. Chem. 2021, 4, 161. [Google Scholar] [CrossRef]
- Smith, J.M.; Chowdhry, R.; Booth, M.J. Controlling Synthetic Cell-Cell Communication. Front. Mol. Biosci. 2022, 8, 809945. [Google Scholar] [CrossRef]
- Llopis-Lorente, A.; Buddingh’, B.C.; Martínez-Máñez, R.; van Hest, J.C.M.; Abdelmohsen, L.K.E. Quorum Sensing Communication between Lipid-Based Artificial Cells. Chem. Commun. 2023, 59, 579–582. [Google Scholar] [CrossRef]
- Smith, J.M.; Hartmann, D.; Booth, M.J. Engineering Cellular Communication between Light-Activated Synthetic Cells and Bacteria. Nat. Chem. Biol. 2023, 19, 1138–1146. [Google Scholar] [CrossRef]
- Gonzales, D.T.; Suraritdechachai, S.; Zechner, C.; Tang, T.-Y.D. Bidirectional Communication between Droplet Interface Bilayers Driven by Cell-Free Quorum Sensing Gene Circuits**. ChemSystemsChem 2023, 5, e202300029. [Google Scholar] [CrossRef]
- Lentini, R.; Martín, N.Y.; Forlin, M.; Belmonte, L.; Fontana, J.; Cornella, M.; Martini, L.; Tamburini, S.; Bentley, W.E.; Jousson, O.; et al. Two-Way Chemical Communication between Artificial and Natural Cells. ACS Cent. Sci. 2017, 3, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Nakano, T. A Biologically Inspired Model of Collective Bio-Nanomachine Rotation via Chemical and Physical Interactions. IEEE Trans. Nanobioscience 2023, 22, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Eto, S.; Matsumura, R.; Shimane, Y.; Fujimi, M.; Berhanu, S.; Kasama, T.; Kuruma, Y. Phospholipid Synthesis inside Phospholipid Membrane Vesicles. Commun. Biol. 2022, 5, 1016. [Google Scholar] [CrossRef] [PubMed]
- Somathilaka, S.S.; Balasubramaniam, S.; Martins, D.P.; Li, X. Revealing Gene Regulation-Based Neural Network Computing in Bacteria. Biophys. Rep. 2023, 3, 100118. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.; Kuscu, M.; Barros, M.T.; Booth, M.; Llopis-Lorente, A.; Magarini, M.; Martins, D.P.; Schäfer, M.; Stano, P. Toward Interdisciplinary Synergies in Molecular Communications: Perspectives from Synthetic Biology, Nanotechnology, Communications Engineering and Philosophy of Science. Life 2023, 13, 208. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, M.; Stano, P.; Egan, M.; Barros, M.T.; Unluturk, B.D.; Payne, G.F. Guest Editorial Special Feature on Bio-Chem-ICTs: Synergies Between Bio/Nanotechnologies and Molecular Communications. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2023, 9, 351–353. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stano, P.; Kuscu, M.; Barros, M.; Egan, M.; Kuruma, Y.; Balasubramaniam, S.; Wang, J.; Nakano, T. Molecular Communication Approaches for Wetware Artificial Life: A Workshop Report. Proceedings 2024, 98, 1. https://doi.org/10.3390/proceedings2024098001
Stano P, Kuscu M, Barros M, Egan M, Kuruma Y, Balasubramaniam S, Wang J, Nakano T. Molecular Communication Approaches for Wetware Artificial Life: A Workshop Report. Proceedings. 2024; 98(1):1. https://doi.org/10.3390/proceedings2024098001
Chicago/Turabian StyleStano, Pasquale, Murat Kuscu, Michael Barros, Malcolm Egan, Yutetsu Kuruma, Sasitharan Balasubramaniam, Jiewen Wang, and Tadashi Nakano. 2024. "Molecular Communication Approaches for Wetware Artificial Life: A Workshop Report" Proceedings 98, no. 1: 1. https://doi.org/10.3390/proceedings2024098001
APA StyleStano, P., Kuscu, M., Barros, M., Egan, M., Kuruma, Y., Balasubramaniam, S., Wang, J., & Nakano, T. (2024). Molecular Communication Approaches for Wetware Artificial Life: A Workshop Report. Proceedings, 98(1), 1. https://doi.org/10.3390/proceedings2024098001







