Abstract
The label-free approach streamlines sample analysis by eliminating the need for fluorescent markers or labels, thus improving accuracy and speed. This contribution explores the potential of silver-based plasmonic gratings as central building blocks for developing biological sensors using label-free detection techniques. It presents the design, fabrication, and optimization of plasmonic gratings, including showcasing their application in biological molecule detection.
1. Introduction
Label-free detection is widely used for studying biological molecules and offers straightforward information compared to using labeled molecules [1]. Silver-based gratings enable label-free detection in biomedical applications by exciting plasmonic surface waves [2]. Additionally, grating structures enable miniaturization, integration with established microelectronic technologies, and simple use in on-chip applications. Plasmonic research focuses on various applications, including chemical and biological sensors, optics, and data transfer. These enable real-time analyte detection through surface plasmon excitation. Grating configurations can be seamlessly combined with integrated circuit manufacturing technology, providing adaptability and accuracy in quantifying small molecule quantities in biosensors. However, the challenges include stability, durability, and fabrication processes. This work presents the development of high-performance biological sensors, harnessing the potential of silver-based plasmonic gratings with microchannel and label-free detection techniques.
2. Materials and Methods
A silver grating was fabricated on a silicon wafer with a surface of (20 × 20) mm2, a thickness of 100 nm, and a grating height of 50 nm. Electron beam lithography was used to create an 800 nm grating period within a total area of 1.2 mm2. A 5 μM solution of protein A [3] in pH 7.5 phosphate-buffered saline (PBS) buffer was measured using polydimethylsiloxane (PDMS) microchannels [4] with a thickness of 2000 μm and a channel height of 150 μm, respectively, attached to the silver grating structure. The grating plasmon excitation was measured with an angle scanning tool. The experimental setup presented in Figure 1 was used to detect the reflected wave from the grating structure, utilizing a He-Ne laser source operated at a wavelength of 632.8 nm. The sample was rotated within an angle range of 10°–45° with reflections analyzed at intervals of 0.2°.
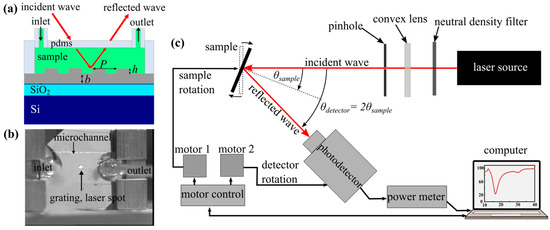
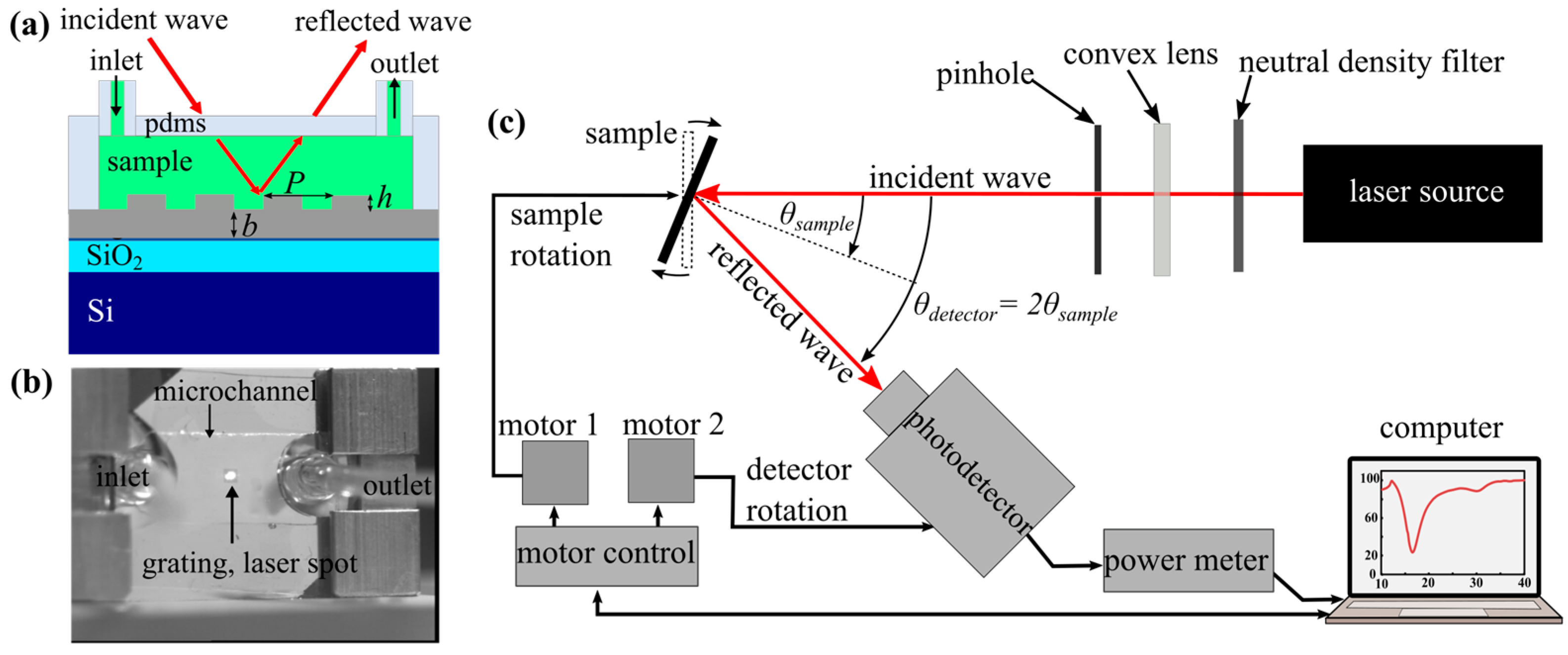
Figure 1.
(a) Schematic overview of the silver grating geometry with a grating period of P = 800 nm, a height of h = 50 nm, and a thickness of b = 100 nm. (b) The silver grating with PDMS microchannel was mounted in the sample holder with laser beam alignment on the grating area. The experimental setup indicating the incident and reflected waves is shown in (c).
3. Discussion
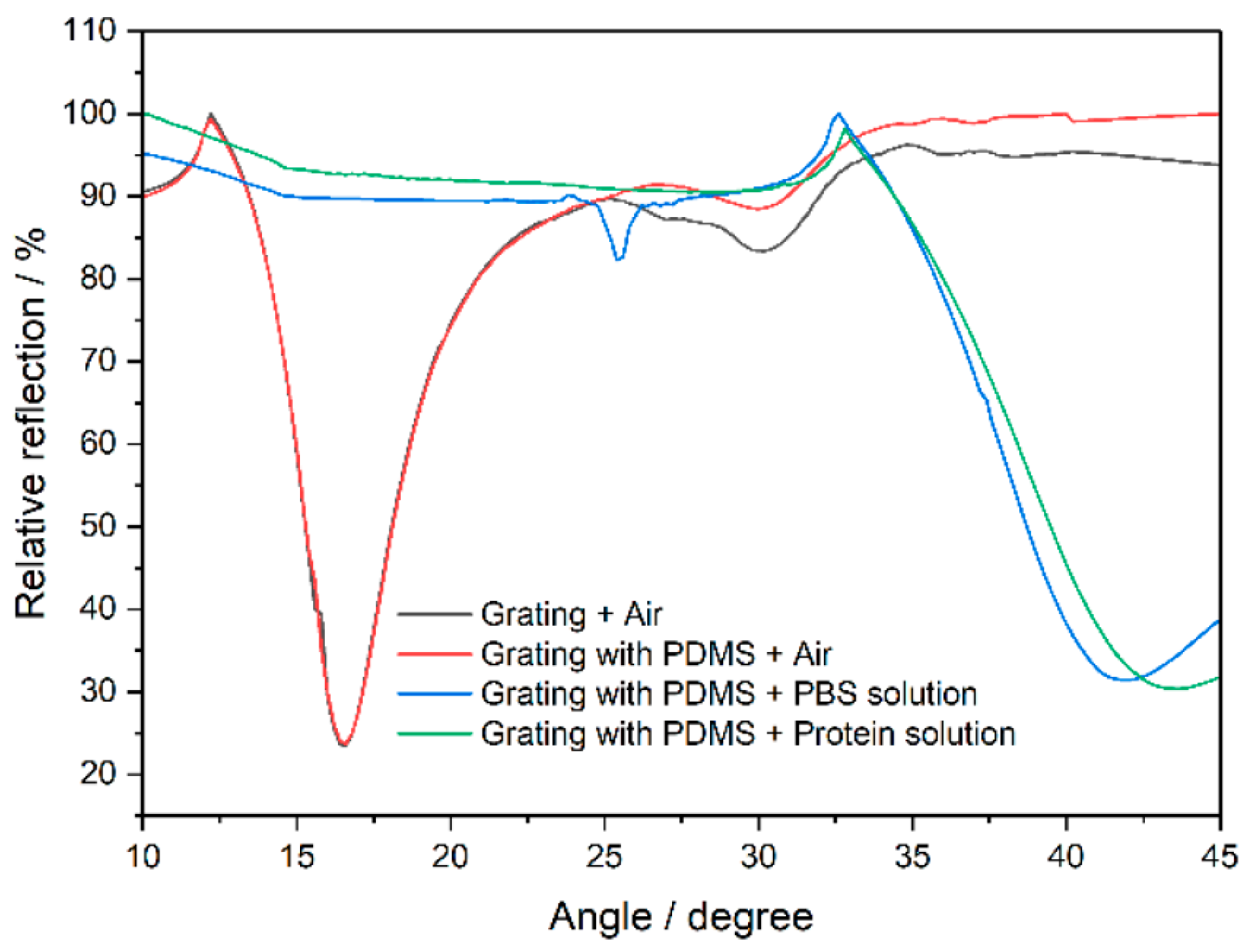
The plasmon excitation angles with and without a PDMS microchannel are depicted in Figure 2 and show similar behavior, indicating its minimal impact on detection. The results confirm PDMS as an appropriate solution-containment channel. The PBS solution shifted the excitation angle of the plasmons, and the protein A solution caused a further shift. The grating sensor with the PDMS microchannel had an average sensitivity of approximately 77.23°/RIU, demonstrating the viability of PDMS as a microchannel and the grating coupler for biomolecular solution identification.
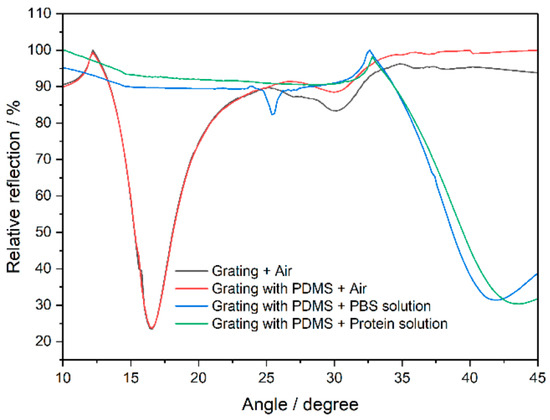
Figure 2.
The effect of PDMS microchannel on the relative reflection and plasmon excitation angle change induced by various fluid solutions.
Author Contributions
Conceptualization by P.S. and D.S.; methodology by P.S.; software by P.S.; writing by S.P. and P.S.; supervision by S.P. All authors have read and agreed to the published version of the manuscript.
Funding
P.S. appreciates funding from the Erasmus Mundus LEADERS (leading mobility between Europe and Asia in developing engineering education and research) program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Cooper, M. (Ed.) Label-Free Biosensors: Techniques and Applications; Label-Free Biosensors; Cambridge University Press: Cambridge, UK, 2009; pp. Xi–Xii. [Google Scholar]
- Li, E.; Chu, H. Plasmonic biosensing devices and systems. In Plasmonic Nanoelectronics and Sensing; EuMA High Frequency Technologies Series; Cambridge University Press: Cambridge, UK, 2014; pp. 217–248. [Google Scholar]
- Christopoulos, T.; Diamandis, E. Immunoassay; Immunoassay Configurations; Academic Press: Cambridge, MA, USA, 1996; pp. 227–236. [Google Scholar]
- Raj, M.K.; Chakraborty, S. PDMS microfluidics: A mini review. J. Appl. Polym. Sci. 2020, 137, 48958. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).