1. Introduction
Diabetes mellitus is a chronic metabolic disorder in which the blood sugar levels are increased abnormally caused by the body’s inability to provide the insulin it needs. Diabetes disease is divided into three different types: type 1 diabetes, type 2 diabetes, and gestational diabetes. In type 1 diabetes, the immune system of the body causes permanent damage to the beta cells in the pancreas. Type 2 diabetes occurs as a result of the body’s inability to use insulin efficiently, and approximately 90% of diabetics have type 2 diabetes [
1]. Last, gestational diabetes occurs when the blood sugar levels are increased during pregnancy.
In a report published by the World Health Organization (WHO) in 2018, it was stated that there are 422 million diabetic patients worldwide [
2]. According to the statistics of the International Diabetes Federation (IDF), this number is expected to reach 642 million people worldwide by 2040. Early diagnosis of diabetes will ensure a healthier treatment of diabetic patients, who are quite high in number, and provides a reduction in risk of some significant diabetes complications such as heart disease, blindness, and renal failure.
Machine learning is frequently preferred in medical diagnosis prediction field because it is capable of discovering important underlying patterns within complex medical data. Classification is one of the most widely applied machine learning tasks which assigns an unknown target value of a new sample to one of the predefined classes. Classification algorithms are widely used in diabetes risk prediction studies [
3,
4,
5,
6,
7,
8,
9]. Sisodia and Singh Sisodia [
3] developed a classification model consists of naive Bayes, decision tree, and Support Vector Machine (SVM) algorithms which predict diabetes risk at an early stage. The experiments in this study were performed on Pima Indians Diabetes Database (PIDD) dataset and the highest accuracy of 76.30% was obtained by naive Bayes algorithm. Kaur and Chhabra [
4] proposed an improved version of J48 decision tree algorithm and tested the improved algorithm on the PIDD dataset. In another study [
5], researchers used an Artificial Neural Network (ANN) method to predict whether a person is diabetics or not. In the experiments, a back-propagation algorithm was utilized for the implementation of the ANN method and the algorithm achieved an 87.3% accuracy rate.
In the literature, there are several hybrid approaches that combine clustering and classification algorithms of machine learning to improve the prediction performance of diabetes [
10,
11,
12,
13]. Chen at al. [
10] developed a hybrid prediction model for type 2 diabetes which merges K-means algorithm with J48 decision tree for data reduction. Their proposed approach gave a high prediction performance with a 90.04% accuracy rate. Researchers in another study [
11] proposed a cloud-based platform with K-means MapReduce to help diagnosis of diabetes. In the experiments, K-means and hierarchical clustering algorithms were compared in terms of performance, running time, and quality. The results indicated that the K-means algorithm processes massive data more efficiently than the hierarchical clustering algorithm.
Nowadays, ensemble learning has been an active paradigm of machine learning which uses multiple learners to improve the prediction performance of the traditional individual methods. Because of this reason, several ensemble learning studies [
14,
15,
16] have been performed on the prediction of diabetes. Mirshahvalad and Zanjani [
14] proposed an ensemble boosting model with a perceptron algorithm to improve the diabetes prediction performance of the traditional perceptron algorithm. Joshi and Alehegn [
15] developed a decision support system using the Adaptive Boosting (AdaBoost) algorithm for the prediction of diabetes mellitus. In this study, a decision stump algorithm was used as a base learner for the AdaBoost algorithm. The proposed system showed an 80.72% accuracy rate compared with SVM, naive Bayes, and decision tree algorithms in the experiments.
Considering these motivations, bagging and boosting approaches using six different decision tree-based (DTB) classifiers (C4.5, random tree, Reduced Error Pruning Tree (REPTree), decision stump, Hoeffding tree, and NBTree) were used on experimental data for the prediction of diabetes at an early stage.
The main contributions of this paper as follows: (i) it presents a brief survey of DTB algorithms and ensemble learning techniques, (ii) it implements bagging and boosting approaches using six different DTB classifiers on diabetes data, and (iii) it compares individual implementation, bagging and boosting of DTB classifiers in terms of accuracy rates.
2. Methods
Classification is the process of categorizing new samples that have no class labels into predefined classes. Considering the relation among the attributes of the training dataset, a new input sample’s unknown class label is predicted. Because classification algorithms provide successful prediction performances, they are preferred in many applications, including document categorization, medical diagnostic prediction, risk assessment, marketing, and sentiment analysis. In this study, six different DTB classification algorithms (C4.5, random tree, REPTree, decision stump, Hoeffding tree, and NBTree) were implemented on the experimental data to predict diabetes risk.
2.1. Decision Tree-Based (DTB) Algorithms
Decision trees are one of the commonly applied classification algorithms that are easy to interpret and create. According to this approach, the classification process is performed by constructing a tree consists of a conjunction of rules. The tree consists of internal nodes, branches, and leaf nodes to represent attributes, attribute values, and classes in the dataset, respectively. In the tree structure, the output of an internal node—namely, branch—is transferred as an input to another internal node.
In the literature, there are several DTB classifiers, including C4.5, random tree, REPTree, decision stump, Hoeffding tree, and NBTree. These algorithms are executed as individuals and base learners for bagging and boosting methods on the diabetes dataset in the experimental study of this paper.
2.1.1. C4.5
C4.5, introduced by Ross Quinlan, is one of the most widely used decision tree algorithms. To construct the tree, first, the information gain values of all attributes in the training set are evaluated and the attribute that shows the highest information gain value is placed in the root node. Then, the tree continues to branch by evaluating information gain value of the remaining attributes. Information gain values of the attributes are calculated as in Equation (1).
where
S is training set,
Sv is a subset of sample space,
A is candidate attribute, and
v is possible values of the attribute.
Entropy value in Equation (1) is calculated as in Equation (2) and indicates the homogeneity of attributes in the dataset.
where
c is the number of classes and
pi is the probability of
ith class.
2.1.2. Random Tree
In random tree algorithms, more than one decision tree is constructed using randomly selected samples from the original dataset. Among all the equal-chanced constructed trees, a random one is selected.
2.1.3. Reduced Error Pruning Tree (REPTree)
REPTree is a rapid decision tree, which generates multiple decision or regression trees using information gain and prunes them using reduced error pruning. Last, it selects the best tree among all generated trees.
2.1.4. Decision Stump
Decision stump, developed by Wayne Iba and Pat Langley, is a one-level DTB algorithm which has only root node and leaves. The algorithm is generally used as a base classifier in boosting ensemble method. In this algorithm, the classification process is performed by considering only one feature in the sample set.
2.1.5. Hoeffding Tree
A Hoeffding tree is a rapid incremental decision tree algorithm for massive data streams because it assumes no change has occurred in the data distribution over time. So, it processes each record in the dataset only once. In this approach, several decision trees are generated and the decision points of these trees are determined by Hoeffding bound.
2.1.6. Naive Bayes Tree (NBTree)
NBTree is a hybrid algorithm that generates a decision tree and applies a naive Bayes classifier at leaves nodes. The aim of this approach is to improve the accuracy of naive Bayes classifiers in larger datasets using a decision tree.
2.2. Ensemble Learning
Ensemble learning is a successful paradigm of machine learning which merges a set of learners instead of using a single learner to predict unknown target attributes. In this structure, all output values obtained from each learner are combined by using a voting mechanism to make final class label prediction. The main goal of ensemble learning is to form a strong classifier consisting of multiple learners to obtain more accurate classification results.
Ensemble learning techniques are mainly group under four categories: bagging, boosting, voting, and stacking. In this study, bagging and boosting approaches which are widely preferred ensemble learning methods are implemented on the experimental data and compared with each other.
2.2.1. Bagging
Bagging, for bootstrap aggregation, is a frequently used ensemble technique that creates multiple training sets using a bootstrap method. In the bootstrap method, multiple training sets are constructed by choosing random and repeatable samples from the original dataset. After creating training subsets, multiple learning models are generated by training each learner in the ensemble structure with these subsets. Last, the predictions obtained from each model are aggregated to obtain the final decision.
Random forest: the random forest algorithm, proposed by Leo Breiman and Adele Cutler, is a bagging algorithm which generates a forest with multiple decision trees. It classifies the unknown target attribute of a new sample by taking a majority vote over all the decision trees in the forest.
2.2.2. Boosting
In the boosting method, the aim is to acquire a strong classifier from a set of weak learners. According to this approach, the samples in the training set are reweighted during the learning phase to generate multiple learners.
AdaBoost: AdaBoost (adaptive boosting), introduced by Yoav Freund and Robert Schapire, is a boosting algorithm that earned Gödel Prize in 2013. In this algorithm, the weight of misclassified samples in the training set is increased in each iteration. Thus, the chance of selecting misclassified samples for the training set is increased and more samples are classified correctly.
MultiBoost: the MultiBoost algorithm is a boosting algorithm that merges wagging (one of the bagging methods), to reduce variance, and the AdaBoost algorithm, for high bias and variance reduction. It uses the C4.5 algorithm as a base learner to generate decision committees.
Real AdaBoost: the real AdaBoost algorithm is another extension of the AdaBoost algorithm introduced by Friedman, Hastie, and Tibshirani. It generates real-valued contributions to the final strong classifier using class probability estimates and applies a linear combination of weak learners.
2.3. Bagging and Boosting Approaches Using DTB Algorithms
In this study, bagging and boosting approaches using DTB algorithms were implemented on the experimental data to predict diabetes risk at an early stage. While bagging and random forest classifiers were selected for the bagging approach, AdaBoost, MultiBoost, and real AdaBoost algorithms were used for the boosting approach of this study. For both bagging (except for random forest algorithm) and boosting approaches, C4.5, random tree, REPTree, decision stump, Hoeffding tree, and NBTree algorithms are selected as base learners. Random forest algorithms do not require any base learner.
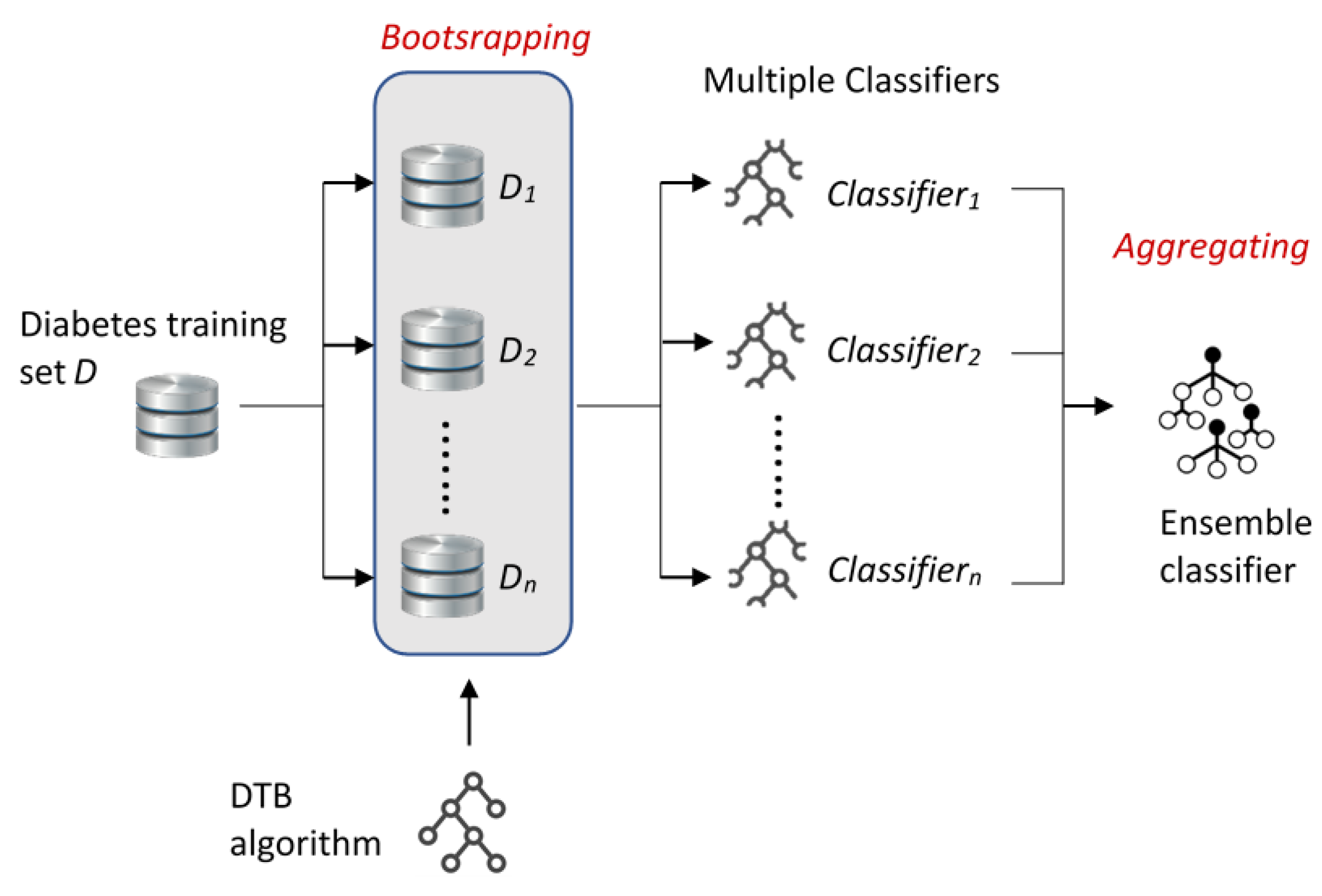
The general structure of bagging and boosting approaches applied in this study is presented in
Figure 1 and
Figure 2, respectively. As shown in
Figure 1, diabetes training dataset
D was divided into
n datasets
D1,
D2, …,
Dn using a bootstrap method. The selected DTB classification algorithm was trained by using these multiple datasets (
D1,
D2, …,
Dn), so multiple classifiers (Classifier
1, Classifier
2, …, Classifier
n) were obtained. Last, the ensemble classifier was constructed by aggregating the multiple classifiers.
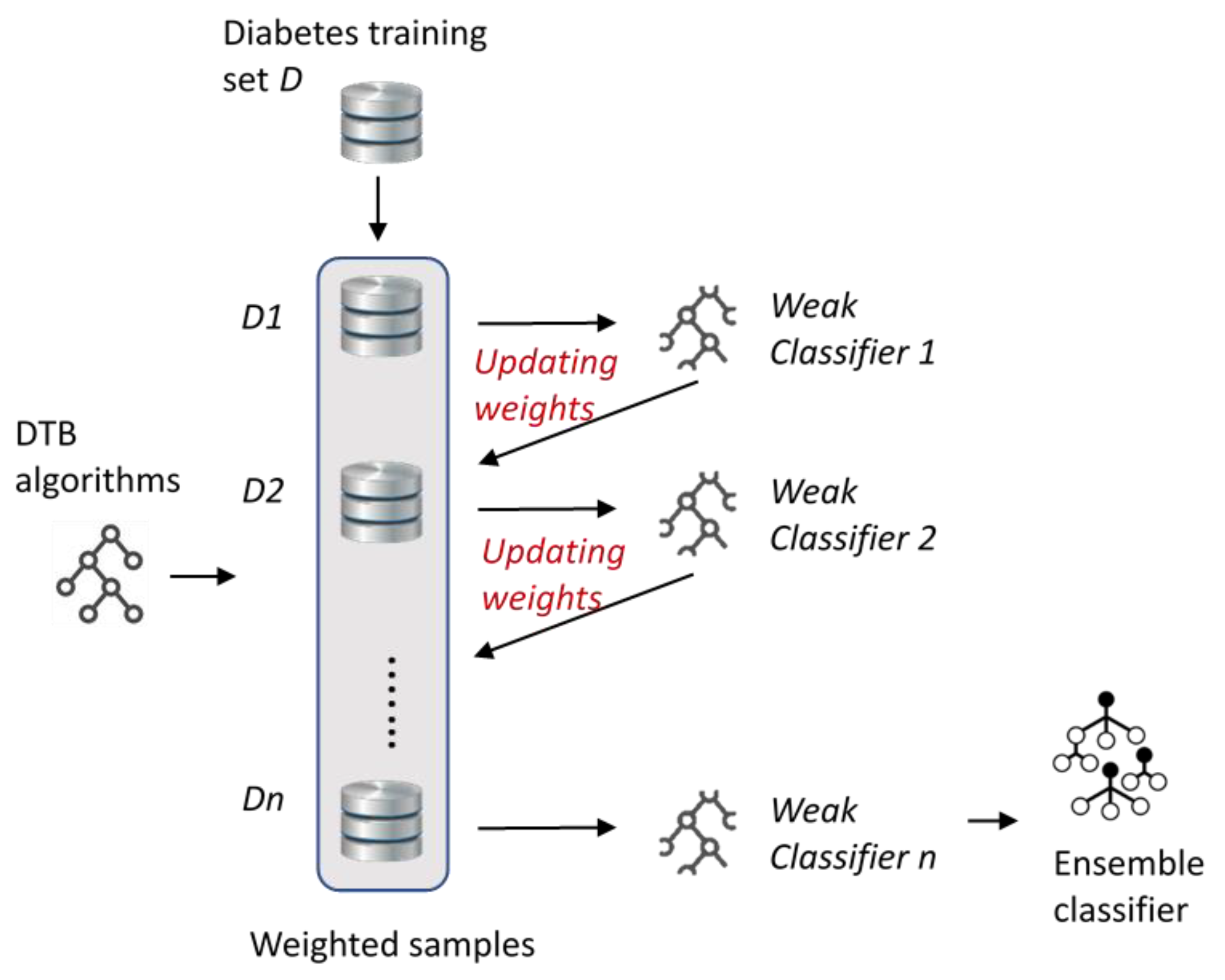
In
Figure 2, the boosting approach of this study is shwon. First, equal-weighted samples were selected from the diabetes training dataset
D to construct the
D1 subset. After this, the selected DTB algorithm was trained with
D1 and the first weak learner was obtained. The samples in the subset were reweighted according to classification accuracies and the new subset (
D2) was constructed. The process of generating weak learners and subsets was repeated until a strong classifier was obtained (ensemble classifier).
3. Experimental Studies
In this experimental study, first, six different DTB classification algorithms (C4.5, random tree, REPTree, decision stump, Hoeffding Tree, and NBTree) were individually implemented on the real-world diabetes dataset to predict diabetes risk at an early stage. Then, bagging (except for the random forest algorithm) and boosting approaches using six different DTB classifiers as base learners were tested on the same dataset. The ensemble approaches (bagging and boosting) were compared with the individual DTB classifiers in terms of accuracy rate. The application in which the experimental studies in this paper were performed was developed by using the Weka open source data mining library [
17].
3.1. Dataset Description
In the experiments, a publicly available diabetes dataset [
18], which consists of diabetes-related symptoms reports, was chosen for demonstrating the capabilities of the ensemble methods with DTB classifiers. The dataset can also be accessed from the data archive in the Statistics Department of University of Florida [
19]. This experimental dataset was constructed through use of a direct questionnaire given to the patients from the Sylhet Diabetes Hospital of Sylhet, Bangladesh. The patients in the questionnaire consist of newly diagnosed diabetic people and nondiabetic people (having some symptoms). The dataset includes 17 attributes and 520 instances. A detailed description of the dataset is shown in
Table 1.
3.2. Results and Discussion
This study presents a comparative classification performance analysis of the application of six different DTB classifiers (C4.5, random tree, REPTree, decision stump, Hoeffding tree, and NBTree) individually and as base learners for bagging and boosting approaches of ensemble learning. In bagging and boosting algorithms, the number of iterations parameter was set as 100. The classification models in this study were tested on the publicly available diabetes dataset and compared to each other in terms of accuracy rate. The accuracy rate values of the models were evaluated by using an n-fold cross validation technique (n = 10).
Accuracy rate is one of the most frequently utilized classification performance measurement methods to present the success of the algorithm on the used dataset. It was evaluated by taking the ratio of the number of correctly classified samples to the total number of samples in the test set, as in Equation (3).
First, six different DTB classification algorithms (C4.5, random tree, REPTree, decision stump, Hoeffding tree, and NBTree) were separately implemented as single classifiers on the diabetes dataset for predicting diabetes risk at an early stage. Afterward, bagging and boosting approaches using these six different DTB classification algorithms as base learners were applied to the same dataset to improve the prediction performance. Bagging and random forest algorithms were used to implement the bagging approach and AdaBoost, MultiBoost, and real AdaBoost algorithms were chosen for the boosting approach of the ensemble structure of this study. Because the random forest algorithm does not require any base learner, it was applied without using a DTB classification algorithm.
Table 2 presents the comparative accuracy rate results of the applied techniques on the experimental dataset. When the individually implemented DTB classifiers’ accuracy rate results are examined, it is clearly seen that the NBTree algorithm gives the best accuracy score of 96.74% among all the classifiers. Additionally, the results indicate that the AdaBoost, MultiBoost, real AdaBoost, and bagging algorithms using DTB classifiers as base learners and the random forest algorithm outperforms the individual DTB classifiers. Additionally, when the experimental results are considered in general, it is observed that NBTree-based real AdaBoost and bagging algorithms provide the most successful diabetes prediction performance with a 98.65% accuracy rate result.
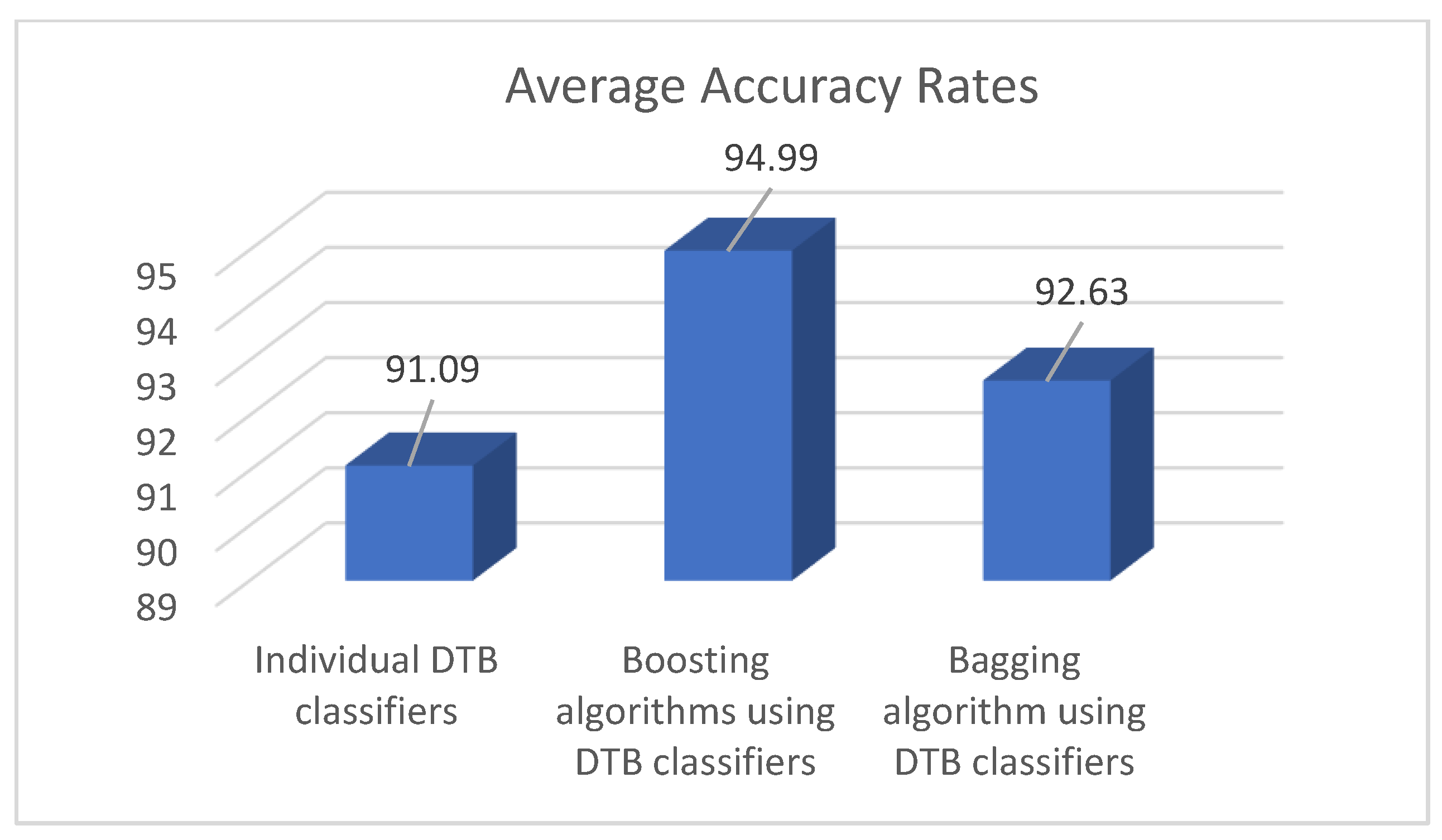
The average accuracy rate of individual DTB classifiers and bagging and boosting approaches using DTB classifiers were calculated and illustrated in the graph given in
Figure 3. The results indicate that the boosting algorithms (AdaBoost, MultiBoost, and Real AdaBoost) using DTB classifiers present the best diabetes prediction performance with an accuracy rate of 94.99%. Additionally, it is possible to say that bagging and boosting approaches using DTB classifiers provide more accurate prediction results than the individual implementation of the DTB classifiers.
4. Conclusions and Future Directions
Diabetes mellitus is a chronic disease that causes an abnormal increase in blood sugar. Early diagnosis of this disease is important for improving the treatment process. Machine learning and ensemble learning techniques provide successful results in the prediction of medical diagnosis field. Thus, in this study, bagging and boosting approaches using six different DTB classifiers (C4.5, random tree, REPTree, decision stump, Hoeffding tree, and NBTree) were implemented on experimental data for the prediction of diabetes at an early stage. In the experiments of this study, individual implementation, bagging, and boosting of DTB classifiers were compared in terms of accuracy rates. When the individually implemented DTB classifiers’ prediction results were examined, it was found that NBTree algorithm presents the highest accuracy score of 96.74% among all the classifiers. Additionally, the results indicate that the bagging and boosting approaches outperform the individual DTB classifiers, and real AdaBoost and bagging algorithms using an NBTree as a base learner gives the best accuracy rate of 98.65%.
For future work, the other ensemble learning types—stacking and voting approaches—using DTB classifiers can be implemented for diabetes prediction. In addition, a hybrid model can be developed by combining the ensemble of clustering methods and classification algorithms to improve the prediction performance of diabetes at an early stage.