Reanalysis of Non-Small-Cell Lung Cancer Microarray Gene Expression Data †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Setup and Visualization of the Dataset
2.3. Gene Expression and Identification of Candidate Genes
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer statistics, 2010. CA Cancer J. Clin. 2010, 60, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, D.P.; Piotrowska, Z.; Lennerz, J.K.; Digumarthy, S.R. Role of imaging biomarkers in mutation-driven non-small cell lung cancer. World J. Clin. Oncol. 2020, 11, 412–427. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zheng, S.; Torossian, A.; Speirs, C.K.; Schleicher, S.; Giacalone, N.J.; Carbone, D.P.; Zhao, Z.; Lu, B. Role of insulin-like growth factor-1 signaling pathway in cisplatin-resistant lung cancer cells. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, e563–e572. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, G.; Di Rienzo, T.; Ojetti, V. Microarray analysis in gastric cancer: A review. World J. Gastroenterol. 2014, 20, 11972–11976. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Khatri, P.; Mazur, P.K.; Polin, M.; Zheng, Y.; Vaka, D.; Hoang, C.D.; Shrager, J.; Xu, Y.; Vicent, S.; et al. A meta-analysis of lung cancer gene expression identifies PTK7 as a survival gene in lung adenocarcinoma. Cancer Res. 2014, 74, 2892–2902. [Google Scholar] [CrossRef]
- Hu, N.; Clifford, R.J.; Yang, H.H.; Wang, C.; Goldstein, A.M.; Ding, T.; Taylor, P.R.; Lee, M.P. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics 2010, 11, 576. [Google Scholar] [CrossRef]
- Bonde, P.; Sui, G.; Dhara, S.; Wang, J.; Broor, A.; Kim, I.F.; Wiley, J.E.; Marti, G.; Duncan, M.; Jaffee, E.; et al. Cytogenetic characterization and gene expression profiling in the rat reflux-induced esophageal tumor model. J. Thorac. Cardiovasc. Surg. 2007, 133, 763–769. [Google Scholar] [CrossRef]
- Arredouani, M.S.; Lu, B.; Bhasin, M.; Eljanne, M.; Yue, W.; Mosquera, J.-M.; Bubley, G.J.; Li, V.; Rubin, M.A.; Libermann, T.A.; et al. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin. Cancer Res. 2009, 15, 5794–5802. [Google Scholar] [CrossRef]
- Bergenfelz, C.; Larsson, A.-M.; von Stedingk, K.; Gruvberger-Saal, S.; Aaltonen, K.; Jansson, S.; Jernström, H.; Janols, H.; Wullt, M.; Bredberg, A.; et al. Systemic Monocytic-MDSCs Are Generated from Monocytes and Correlate with Disease Progression in Breast Cancer Patients. PLoS ONE 2015, 10, e0127028. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, L.-H.; Wang, X.-H.; Xing, X.-F.; Cheng, X.-J.; Dong, B.; Hu, Y.; Du, H.; Li, Y.-A.; Zhu, Y.-B.; et al. PTK7 as a novel marker for favorable gastric cancer patient survival. J. Surg. Oncol. 2012, 106, 880–886. [Google Scholar] [CrossRef]
- Lu, T.-P.; Tsai, M.-H.; Lee, J.-M.; Hsu, C.-P.; Chen, P.-C.; Lin, C.-W.; Shih, J.-Y.; Yang, P.-C.; Hsiao, C.K.; Lai, L.-C.; et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2590–2597. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.-P.; Hsiao, C.K.; Lai, L.-C.; Tsai, M.-H.; Hsu, C.-P.; Lee, J.-M.; Chuang, E.Y. Identification of regulatory SNPs associated with genetic modifications in lung adenocarcinoma. BMC Res. Notes 2015, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Uriarte, R.; Alvarez de Andrés, S. Gene selection and classification of microarray data using random forest. BMC Bioinform. 2006, 7, 3. [Google Scholar] [CrossRef]
- Huang, H.-L.; Chang, F.-L. ESVM: Evolutionary support vector machine for automatic feature selection and classification of microarray data. Biosystems 2007, 90, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-M.; Yao, C.-T.; Chang, Y.-T.; Chou, H.-L.; Chou, Y.-C.; Chen, K.-H.; Terng, H.-J.; Huang, C.-S.; Lee, C.-C.; Su, S.-L.; et al. Gene Expression Profiling of Colorectal Tumors and Normal Mucosa by Microarrays Meta-Analysis Using Prediction Analysis of Microarray, Artificial Neural Network, Classification, and Regression Trees. Available online: https://www.hindawi.com/journals/dm/2014/634123/ (accessed on 2 September 2020).
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.; Wong, C.-J. Working with Bioconductor Objects: Microarray Analysis; Fred Hutchinson Cancer Research Center. 2010. Available online: https://www.bioconductor.org/help/course-materials/2011/intl-workshop-bioc/presentation-slides/Introduction-Lab.pdf (accessed on 17 November 2020).
- Gentleman, R.; Carey, V.; Huber, W.; Hahne, F. Genefilter: Genefilter: Methods for Filtering Genes from High-Throughput Experiments; Bioconductor Version: Release (3.12); Bioconductor 2020. Available online: https://bioconductor.org/packages/release/bioc/html/genefilter.html (accessed on 16 November 2020).
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Kuhn, M. The Caret Package; R Foundation for Statistical Computing: Vienna, Austria, 2012; Available online: https://cran.r-project.org/package=caret (accessed on 16 November 2020).
- Chen, S.; Li, J.; Zhou, P.; Zhi, X. SPTBN1 and cancer, which links? J. Cell. Physiol. 2020, 235, 17–25. [Google Scholar] [CrossRef]
- Huang, H.; Li, T.; Ye, G.; Zhao, L.; Zhang, Z.; Mo, D.; Wang, Y.; Zhang, C.; Deng, H.; Li, G.; et al. High expression of COL10A1 is associated with poor prognosis in colorectal cancer. Onco Targets Ther. 2018, 11, 1571–1581. [Google Scholar] [CrossRef]
- Ahn, N.; Kim, W.-J.; Kim, N.; Park, H.W.; Lee, S.-W.; Yoo, J.-Y. The Interferon-Inducible Proteoglycan Testican-2/SPOCK2 Functions as a Protective Barrier against Virus Infection of Lung Epithelial Cells. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Liao, Y.-X.; Zeng, J.-M.; Zhou, J.-J.; Yang, G.-H.; Ding, K.; Zhang, X.-J. Silencing of RTKN2 by siRNA suppresses proliferation, and induces G1 arrest and apoptosis in human bladder cancer cells. Mol. Med. Rep. 2016, 13, 4872–4878. [Google Scholar] [CrossRef]
- Zhang, W.; Fan, J.; Chen, Q.; Lei, C.; Qiao, B.; Liu, Q. SPP1 and AGER as potential prognostic biomarkers for lung adenocarcinoma. Oncol. Lett. 2018, 15, 7028–7036. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhu, W.; Xiao, G.; Ding, M.; Chang, J.; Liao, H. Effect of AGER on the biological behavior of non-small cell lung cancer H1299 cells. Mol. Med. Rep. 2020, 22, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ma, H.; Deng, G.; Tang, L.; Lu, J.; Chen, X. Clinical significance and biological function of fucosyltransferase 2 in lung adenocarcinoma. Oncotarget 2017, 8, 97246–97259. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, Z.; Shen, W.; Li, X.; Dhoomun, D.K.; Tian, Y. Golgi Membrane Protein 1 (GOLM1) Promotes Growth and Metastasis of Breast Cancer Cells via Regulating Matrix Metalloproteinase-13 (MMP13). Med. Sci. Monit. 2019, 25, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Han, H.-B.; Gu, J.; Zuo, H.-J.; Chen, Z.-G.; Zhao, W.; Li, M.; Ji, D.-B.; Lu, Y.-Y.; Zhang, Z.-Q. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J. Pathol. 2012, 226, 544–555. [Google Scholar] [CrossRef]
- Sauter, W.; Rosenberger, A.; Beckmann, L.; Kropp, S.; Mittelstrass, K.; Timofeeva, M.; Wölke, G.; Steinwachs, A.; Scheiner, D.; Meese, E.; et al. Matrix Metalloproteinase 1 (MMP1) Is Associated with Early-Onset Lung Cancer. Cancer Epidemiol. Biomark. Prev. 2008, 17, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
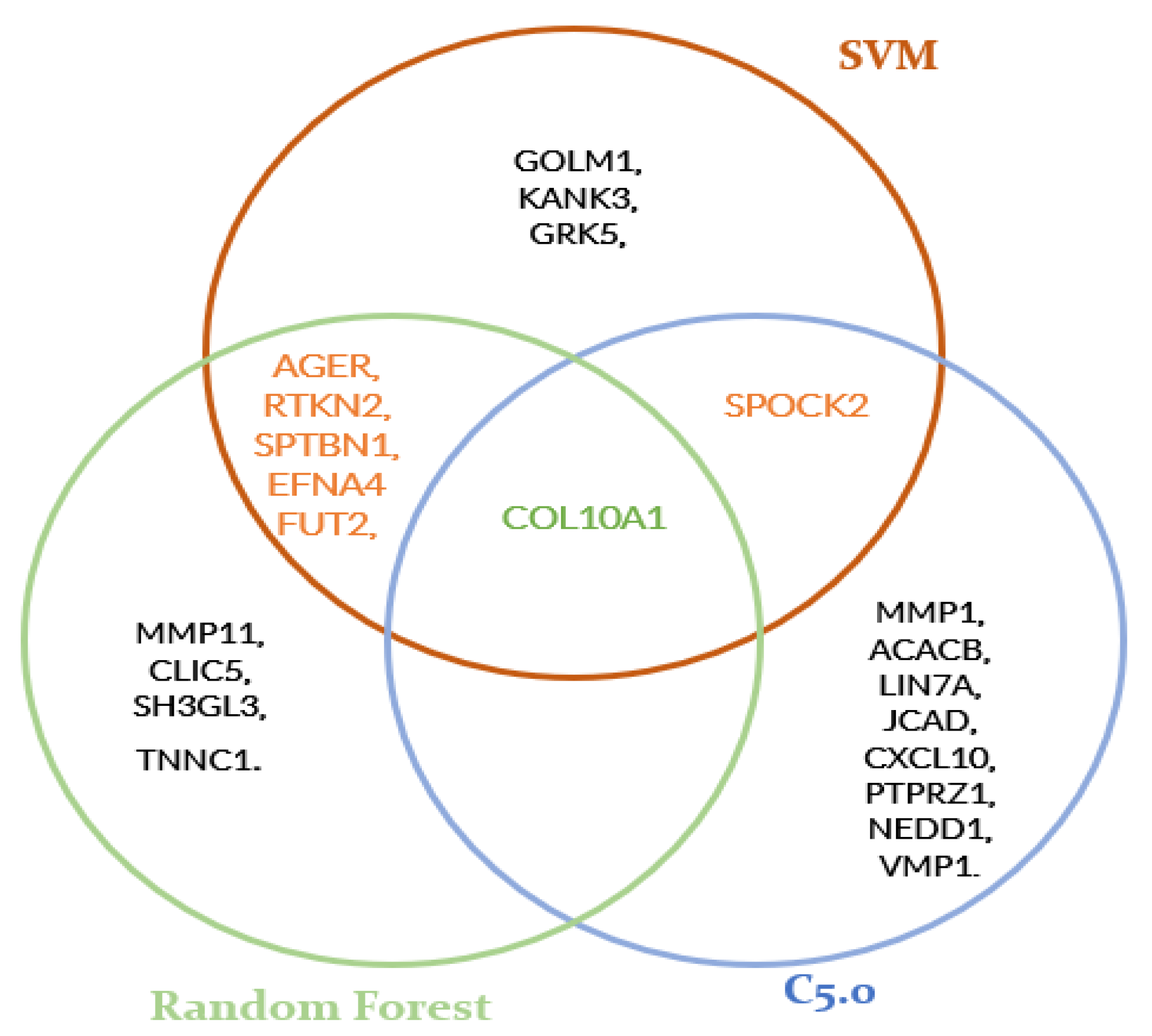
| PROB_ID | SYMBOL | GENE | |
|---|---|---|---|
| 1 | 217428_s_at | COL10A1 | collagen type X alpha 1 chain |
| 2 | 203878_s_at | MMP11 | matrix metallopeptidase 11 |
| 3 | 213317_at | CLIC5 | chloride intracellular channel 5 |
| 4 | 210608_s_at | FUT2 | fucosyltransferase 2 |
| 5 | 215918_s_at | SPTBN1 | spectrin beta, non-erythrocytic 1 |
| 6 | 205637_s_at | SH3GL3 | SH3 domain containing GRB2 like 3, endophilin A3 |
| 7 | 205107_s_at | EFNA4 | ephrin A4 |
| 8 | 230469_at | RTKN2 | rhotekin 2 |
| 9 | 210081_at | AGER | advanced glycosylation end-product specific receptor |
| 10 | 209904_at | TNNC1 | troponin C1, slow skeletal and cardiac type |
| PROB_ID | SYMBOL | GENE | |
|---|---|---|---|
| 1 | 202524_s_at | SPOCK2 | SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| 2 | 217771_at | GOLM1 | golgi membrane protein 1 |
| 3 | 230469_at | RTKN2 | rhotekin 2 |
| 4 | 215918_s_at | SPTBN1 | spectrin beta, non-erythrocytic 1 |
| 5 | 213715_s_at | KANK3 | KN motif and ankyrin repeat domains 3 |
| 6 | 217428_s_at | COL10A1 | collagen type X alpha 1 chain |
| 7 | 204396_s_at | GRK5 | G protein-coupled receptor kinase 5 |
| 8 | 205107_s_at | EFNA4 | ephrin A4 |
| 9 | 210608_s_at | FUT2 | fucosyltransferase 2 |
| 10 | 210081_at | AGER | advanced glycosylation end-product specific receptor |
| PROB_ID | SYMBOL | GENE | |
|---|---|---|---|
| 1 | 202524_s_at | SPOCK2 | SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| 2 | 217428_s_at | COL10A1 | collagen type X alpha 1 chain |
| 3 | 49452_at | ACACB | acetyl-CoA carboxylase beta |
| 4 | 227929_at | LIN7A | lin-7 homolog A, crumbs cell polarity complex component |
| 5 | 213316_at | JCAD | junctional cadherin 5 associated |
| 6 | 204533_at | CXCL10 | C-X-C motif chemokine ligand 10 |
| 7 | 204469_at | PTPRZ1 | protein tyrosine phosphatase receptor type Z1 |
| 8 | 1552417_a_at | NEDD1 | NEDD1 gamma-tubulin ring complex targeting factor |
| 9 | 1569003_at | VMP1 | vacuole membrane protein 1 |
| 10 | 204475_at | MMP1 | matrix metallopeptidase 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bawa, T.A.; Özkan, Y.; Erol, Ç.S. Reanalysis of Non-Small-Cell Lung Cancer Microarray Gene Expression Data. Proceedings 2021, 74, 22. https://doi.org/10.3390/proceedings2021074022
Bawa TA, Özkan Y, Erol ÇS. Reanalysis of Non-Small-Cell Lung Cancer Microarray Gene Expression Data. Proceedings. 2021; 74(1):22. https://doi.org/10.3390/proceedings2021074022
Chicago/Turabian StyleBawa, Tcharé Adnaane, Yalçın Özkan, and Çiğdem Selçukcan Erol. 2021. "Reanalysis of Non-Small-Cell Lung Cancer Microarray Gene Expression Data" Proceedings 74, no. 1: 22. https://doi.org/10.3390/proceedings2021074022
APA StyleBawa, T. A., Özkan, Y., & Erol, Ç. S. (2021). Reanalysis of Non-Small-Cell Lung Cancer Microarray Gene Expression Data. Proceedings, 74(1), 22. https://doi.org/10.3390/proceedings2021074022





