Temporal Changes and Alternating Host Tree Root and Shoot Growth Affect Soil Microbiomes †
Abstract
:1. Introduction
2. Materials and Methods
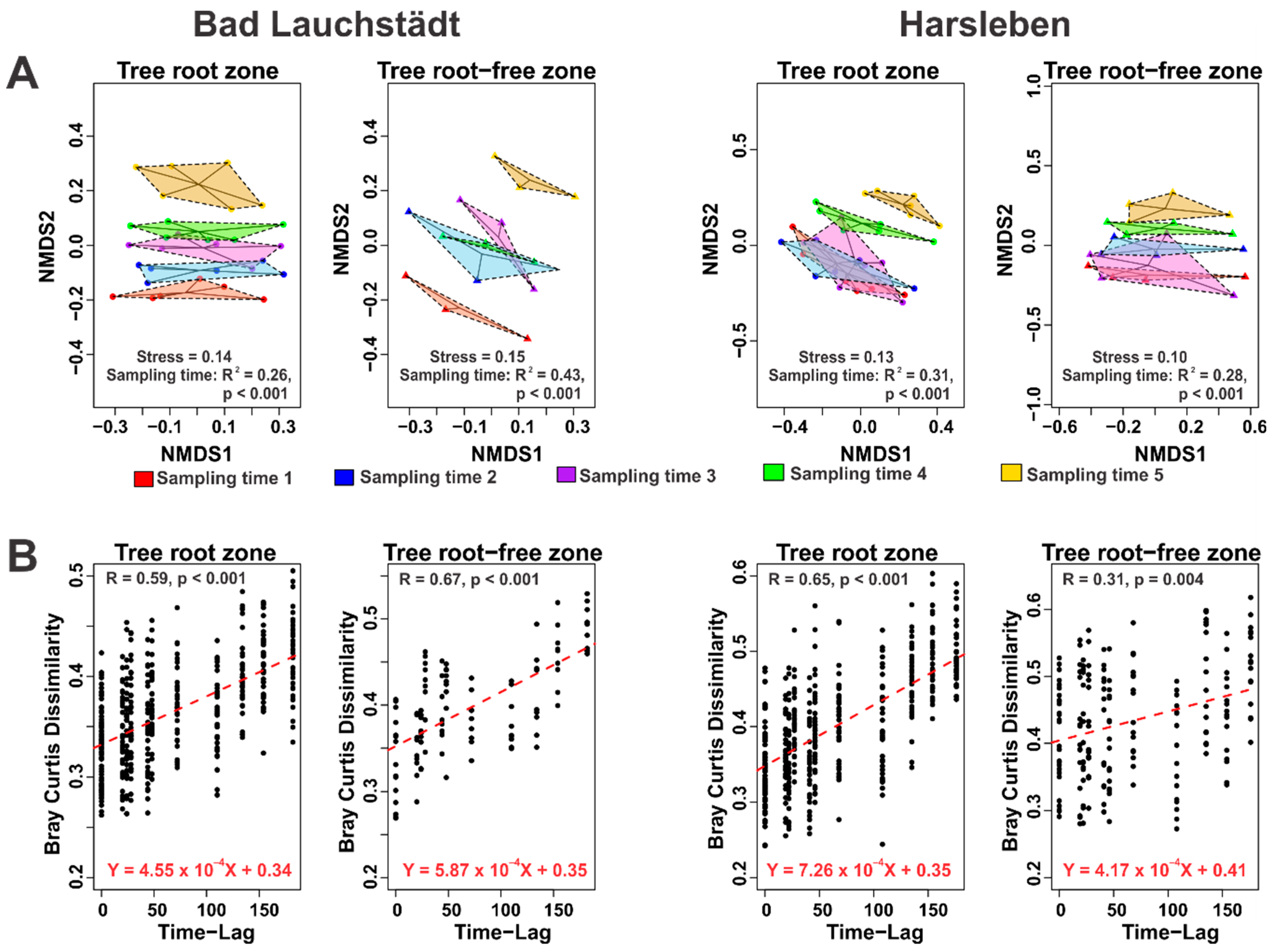
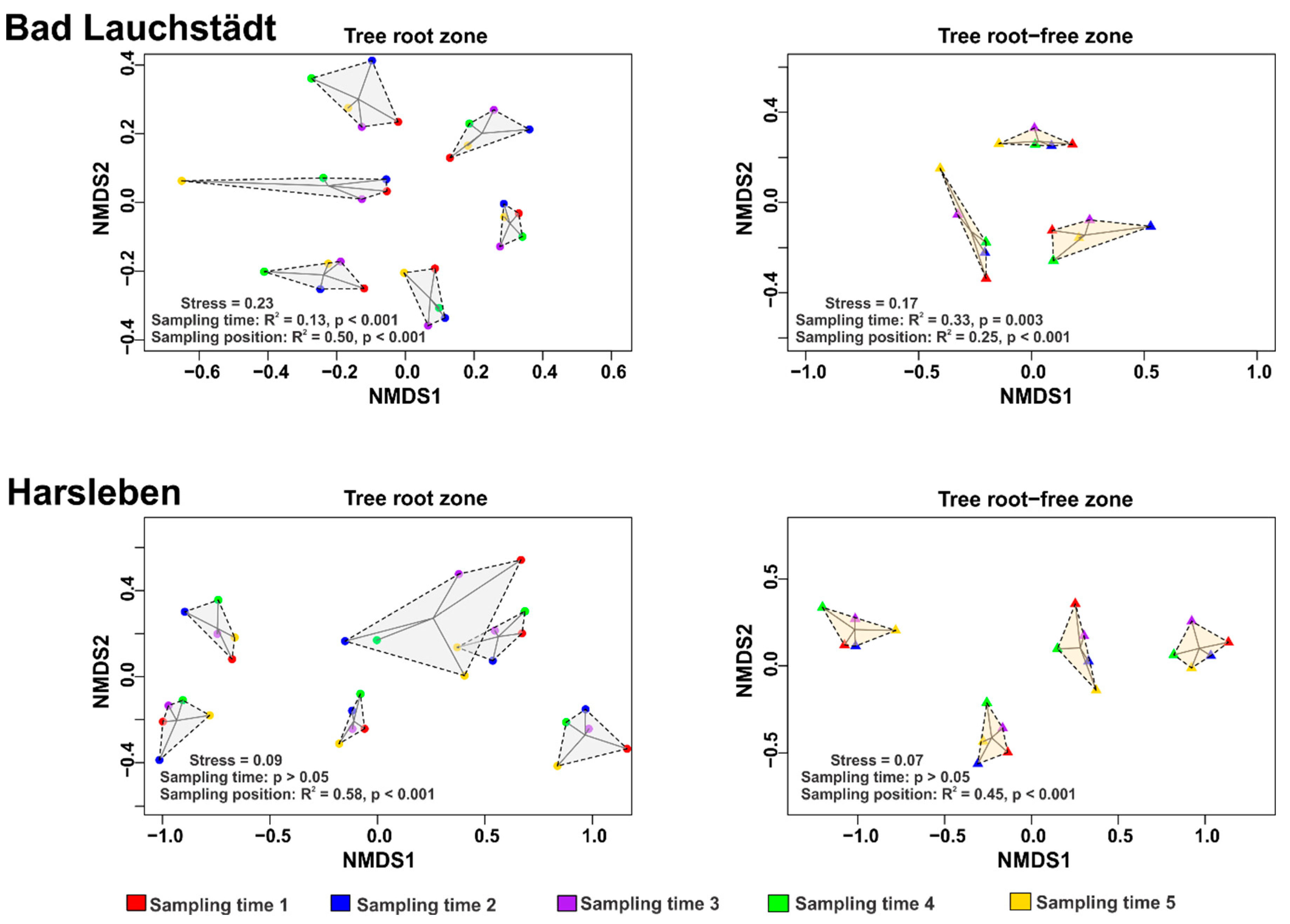
3. Results and Discussion
4. Conclusions
Acknowledgments
References
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef]
- Aulakh, M.; Wassmann, R.; Bueno, C.; Kreuzwieser, J.; Rennenberg, H. Characterization of root exudates at different growth stages of ten rice (Oryza sativa L.) cultivars. Plant Biol. 2001, 3, 139–148. [Google Scholar] [CrossRef]
- Hannula, S.E.; Kielak, A.M.; Steinauer, K.; Huberty, M.; Jongen, R.; De Long, J.R.; Heinen, R.; Bezemer, T.M. Time after Time: Temporal Variation in the Effects of Grass and Forb Species on Soil Bacterial and Fungal Communities. mBio 2019, 10, e02635-19. [Google Scholar] [CrossRef]
- Herrmann, S.; Recht, S.; Boenn, M.; Feldhahn, L.; Angay, O.; Fleischmann, F.; Tarkka, M.T.; Grams, T.E.E.; Buscot, F. Endogenous rhythmic growth in oak trees is regulated by internal clocks rather than resource availability. J. Exp. Bot. 2015, 66, 7113–7127. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, S.; Grams, T.E.E.; Tarkka, M.T.; Angay, O.; Bacht, M.; Bönn, M.; Feldhahn, L.; Graf, M.; Kurth, F.; Maboreke, H.; et al. Endogenous rhythmic growth, a trait suitable for the study of interplays between multitrophic interactions and tree development. Perspect. Plant Ecol. Evol. Syst. 2016, 19, 40–48. [Google Scholar] [CrossRef]
- Angay, O.; Fleischmann, F.; Recht, S.; Herrmann, S.; Matyssek, R.; Oßwald, W.; Buscot, F.; Grams, T.E.E. Sweets for the foe—Effects of nonstructural carbohydrates on the susceptibility of Quercus robur against Phytophthora quercina. New Phytol. 2014, 203, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Karst, J.; Gaster, J.; Wiley, E.; Landhäusser, S.M. Stress differentially causes roots of tree seedlings to exude carbon. Tree Physiol. 2016, 37, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Denef, K.; Roobroeck, D.; Manimel Wadu, M.C.W.; Lootens, P.; Boeckx, P. Microbial community composition and rhizodeposit-carbon assimilation in differently managed temperate grassland soils. Soil Biol. Biochem. 2009, 41, 144–153. [Google Scholar] [CrossRef]
- Habiyaremye, J.D.D.; Goldmann, K.; Reitz, T.; Herrmann, S.; Buscot, F. Tree root zone microbiome: exploring the magnitude of environmental conditions and host tree impact. Front. Microbiol. 2020, 11, 749. [Google Scholar] [CrossRef] [PubMed]
- Weißbecker, C.; Schnabel, B.; Heintz-Buschart, A. Dadasnake, a Snakemake implementation of DADA2 to process amplicon sequencing data for microbial ecology. GigaScience 2020, 9, giaa135. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Köster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2019. Available online: https://www.r-project.org (accessed on 17 August 2020).
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; et al. vegan: Community Ecology Package. R package Version 2.5-6; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Sun, S.; Li, S.; Avera, B.N.; Strahm, B.D.; Badgley, B.D. Soil Bacterial and Fungal Communities Show Distinct Recovery Patterns during Forest Ecosystem Restoration. Appl. Environ. Microbiol. 2017, 83, e00966-17. [Google Scholar] [CrossRef]
- Lauber, C.L.; Ramirez, K.S.; Aanderud, Z.; Lennon, J.; Fierer, N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013, 7, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Burke, D.J.; López-Gutiérrez, J.C.; Smemo, K.A.; Chan, C.R. Vegetation and Soil Environment Influence the Spatial Distribution of Root-Associated Fungi in a Mature Beech-Maple Forest. Appl. Environ. Microbiol. 2009, 75, 7639–7648. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habiyaremye, J.d.D.; Herrmann, S.; Buscot, F.; Goldmann, K. Temporal Changes and Alternating Host Tree Root and Shoot Growth Affect Soil Microbiomes. Proceedings 2020, 66, 35. https://doi.org/10.3390/proceedings2020066035
Habiyaremye JdD, Herrmann S, Buscot F, Goldmann K. Temporal Changes and Alternating Host Tree Root and Shoot Growth Affect Soil Microbiomes. Proceedings. 2020; 66(1):35. https://doi.org/10.3390/proceedings2020066035
Chicago/Turabian StyleHabiyaremye, Jean de Dieu, Sylvie Herrmann, François Buscot, and Kezia Goldmann. 2020. "Temporal Changes and Alternating Host Tree Root and Shoot Growth Affect Soil Microbiomes" Proceedings 66, no. 1: 35. https://doi.org/10.3390/proceedings2020066035
APA StyleHabiyaremye, J. d. D., Herrmann, S., Buscot, F., & Goldmann, K. (2020). Temporal Changes and Alternating Host Tree Root and Shoot Growth Affect Soil Microbiomes. Proceedings, 66(1), 35. https://doi.org/10.3390/proceedings2020066035




