Abstract
Bacteria of the Rhizobium genus form a group of microorganisms existing in the environment in two forms: symbiotic, in the root nodules of Fabaceae sp. plants and free-living, and saprophytic in the soil environment. The subject of this study was genetic identification and characterization of metabolic activity of different strains from Rhizobium genus bacteria. The study was conducted on the 16 bacteria strains from the collection of the Department of Agricultural Microbiology, Institute of Soil Science and Plant Cultivation in Puławy, Poland. Based on the sequencing of PCR products, we found that all strains belong to one species—Rhizobium leguminosarum. The study of metabolic activity was performed using the GEN III BIOLOG system method (Biolog Inc., Hayward, CA, USA). Metabolism analysis of all R. leguminosarum strains with the use of GEN III™ plates showed that carbohydrates (CH) were the most intensively utilised group of substrates. Between the Rhizobium leguminosarum strains, there are metabolic differences in terms of the studied features.
1. Introduction
Bacteria of the Rhizobium genus form a group of microorganisms existing in the environment in two forms: symbiotic, in the root nodules of Fabaceae sp. plants and free-living, and saprophytic in the soil environment [1]. Inside of the root, these bacteria differentiate into nitrogen-fixing bacteroids [2]. The basic function of Rhizobium sp. in a symbiosis is to reduce nitrogen to ammonia directly assimilated by the plant. Nitrogen reduction occurs with the participation of enzymatic nitrogenase complex [3]. The subject of study was genetic identification and characterization of metabolic activity of different strains from Rhizobium genus.
2. Material and Methods
2.1. Bacterial Strains
The study was conducted on the 16 bacteria strains from the collection of Department of Agricultural Microbiology, Institute of Soil Science and Plant Cultivation in Puławy, Poland. Bacteria strains were isolated from root nodules derived from plans of the genus Trifolium. The collection of bacterial strains was obtained from the roots of plants growing in many locations, mainly in Poland (Table 1).

Table 1.
Source of bacterial strains.
2.2. PCR and Sequencing
For PCR, a small amount of material was taken from a single bacterial colony and transferred to a sterile eppendorf with 20 µL of MiliQ water. The samples were thoroughly mixed, and 1 µL was taken from the mixture for the PCR reaction. The 16S rDNA region was amplified using primers: 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT) [4]. The PCR products were sequenced in Genomed S.A (Warsaw, Poland) using the same primers as at the PCR step. The sequences from both primers were assembled in Unipro UGENE 1.25 software [5], and the Blast search was performed [6].
2.3. Biolog GEN III
The study of metabolic activity was performed using the GEN III BIOLOG system method (Biolog Inc., Hayward, CA, USA). The GEN III microplate contains 94 phenotypic tests: 71 carbon source utilization assays and 23 chemical sensitivity assays. Tetrazolium dyes from the wells of the microplate were used to indicate the use of carbon sources or resistance to inhibitory chemicals by microorganisms. The cell suspensions were inoculated into the 134 GEN III™ (100 μL per well) and incubated at 25 °C for 7 days. The intensity of colour development was recorded at λ = 590 nm at 24 h intervals for a period of 168 h. The results obtained at 168 h are presented because the most intensive substrate decomposition was observed after this incubation time.
2.4. Data Analysis and Visualisation
Heatmaps were generated using GenIII Omnilog values (data after 168 h of incubation) with R software (version 3.5.1, Northern Ave, Boston, MA, USA) and pheatmap package. Similarity trees were constructed using Bray–Curtis cluster analysis with the UPGMA method [7].
Statistical analyses were performed using the packet Statistica.PL ver. 10.0 (StatSoft. Inc., Tulsa, OK, USA). The dendrogram applied Ward’s method clustering and the squared Euclidean distance matrix calculation method [8].
3. Results
3.1. Bacterial Species
PCR products from all the isolates were compared with a size marker, and a product with an expected size of about 1500 bp was found (Figure 1). Based on the sequencing of PCR products, we found that all strains belong to one species—Rhizobium leguminosarum with a sequence identity of 97–100% (NCBI GenBank, Table 2) [6].
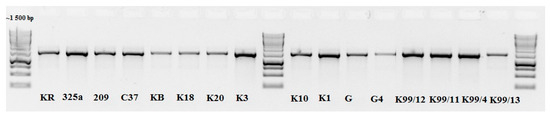
Figure 1.
Picture of polyacrylamide gel electrophoresis.

Table 2.
The identification of bacteria strains (NCBI GenBank).
3.2. Metabolic Activity of Rhizobium Leguminosarum Bacteria
Based on results, the heat maps were made (Figure 2) and the cluster analysis according to Ward’s method conducted, thus illustrating the diversity of strains in terms of the intensity and pace of the consumption of the individual compounds (Figure 3).
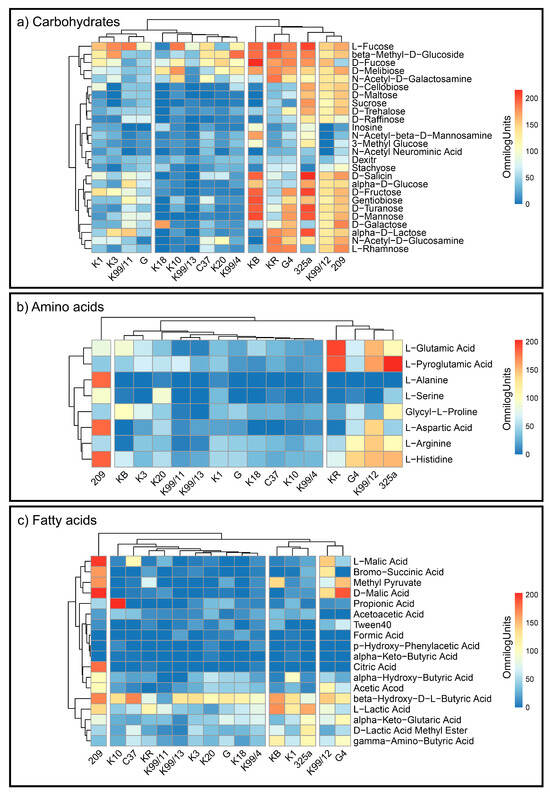
Figure 2.
Heatmaps for the carbon utilization patterns of the substrates GEN III grouped into three biochemical groups: (a) carbohydrates, (b) amino acids, (c) fatty acids, by each strain of Rhizobium leguminosarum. Data are shown after 168 h of incubation. The gradient from light blue to red represents positive utilization.
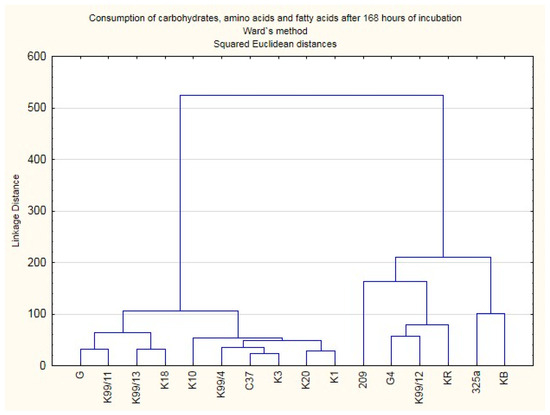
Figure 3.
Dendrogram showing division of Rhizobium leguminosarum strains due to the use of carbohydrates, amino acids, and fatty acids as a carbon sources after 168 h incubation.
Metabolism analysis of all R. leguminosarum strains with the use of GEN III™ plates showed that carbohydrates (CH) were the most intensively utilised group of substrates. Between the Rhizobium leguminosarum strains, there were metabolic differences in terms of the studied features (Figure 2). This may indicate the adaptive capacity of microorganisms to the environmental conditions in which they currently live. Based on the cluster analysis, 3 groups of microorganisms were isolated in terms of the intensity of decomposition of the tested compounds (Figure 3). The most active strains in terms of use of all types of carbon substrates were strains 209, K99/12, 325a and G4.
4. Conclusions
In conclusion, it can be stated that, between the Rhizobium leguminosarum strains, there are metabolic differences in terms of the studied features. This may indicate the adaptive capacity of microorganisms to the environmental conditions in which they currently live.
Author Contributions
K.G. and A.G. conceived and designed the experiments; K.G., K.F. and J.G. performed the experiments and analysed the data; K.G. and K.F. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
The research was partially funded by the Ministry of Science and Higher Education, research task: “Determination of the effect of co-inoculation of clover (Trifolium pretense L.) with Azospirillum spp. and Rhizobium spp. for the growth and nodulation of plants under conditions of contamination by polycyclic aromatic hydrocarbons”/2016 and the frames of Task 1.4. Multi—Annual Programme IUNG—PIB (2016–2020).
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Stasiak, G.; Mazur, A.; Koper, P.; Żebracki, K.; Skorupska, A. Symbiosis of rhizobia with legume plants (Fabaceae). Postep. Mikrobiol. 2016, 55, 289–299. [Google Scholar]
- Kereszt, A.; Mergaert, P.; Kondorosi, E. Bacteroid development in legume nodules: Evolution of mutual benefit or of sacrificial victims? Mol. Plant-Microbe Interact. 2011, 24, 1300–1309. [Google Scholar] [CrossRef]
- Oke, V.; Long, S.R. Bacteroid formation in the Rhizobium-legume symbiosis. Curr. Opin. Microbiol. 1999, 2, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. UGENE team Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Blast NCBI. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 October 2019).
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).