Abstract
The enterobacteria that produce β-lactamases are the main focus of infections in the healthcare environment. This is due to the difficulty they present in terms of treatment, their ease of transmission, and the impact they represent at the economic and personal level. The bacteria of greatest clinical relevance are those with resistance to third and fourth generation cephalosporins, extended spectrum β-lactamase and AmpC. Currently, carbapenemics are one of the few antimicrobials effective against multi-drug resistant organisms. However, the emergence of carbapenem-resistant enterobacteria has increased health concerns. These microorganisms include K. pneumoniae, a pan-resistant bacteria with high morbidity and mortality rates in public health facilities. In this work, we have carried out a review on the antimicrobial resistance genes found in its genome, as well as the resistance mechanisms involved. Finally, we will focus on the main outbreaks causing nosocomial infections during the last few years.
1. Introduction
Klebsiella pneumoniae is an encapsulated Gram-negative bacillus belonging to the Enterobacteriaceae family. It was first described in 1882 by Carl Friedlander, from lung tissues of dead pneumonia patients. It presents a mucoid phenotype in media such as MacConkey or eosin methylene blue agars. Moreover, it is capable of fermenting lactose and mannitol, and reducing nitrate to nitrite. K. pneumoniae is a bacterium able to infect humans, causing different types of infections, including respiratory and urinary tract infections, soft tissue infections, surgical wounds and sepsis. After E. coli, K. pneumoniae is the second most important opportunistic enterobacterium causing nosocomial and community infections. However, it is a ubiquitous species in nature, although environmental strains are more susceptible to antibiotics than clinical ones. It is a consequence of the selection pressure exerted by antibiotics in health care settings [1].
The pangenome of K. pneumoniae (total set of genes) is approximately 5.5 Mbp in size and codes for an average of 5500 genes. More than 30,000 unique sequences have been identified that code for different proteins. However, it is still an open pangenome, in which there are accessory genes that have not yet been characterized [1,2]. Its core genome (set of genes shared by all members of the species) is composed of approximately 2000 genes. These genes have been determined by examining their presence in more than 95% of the isolates from that species [1,2]. The accessory genome contains genes that vary between bacteria of the same species. This includes both chromosomally encoded genes and those located in the mobile genetic elements. In K. pneumoniae, most of the genome is made up of accessory genes. This allows their classification into different strains, and different pathotypes can be catalogued in the clinical setting [opportunistic, Carbapenem-Resistant Enterobacteria (CRE) or hypervirulent (hv) bacteria] (Figure 1) [1].
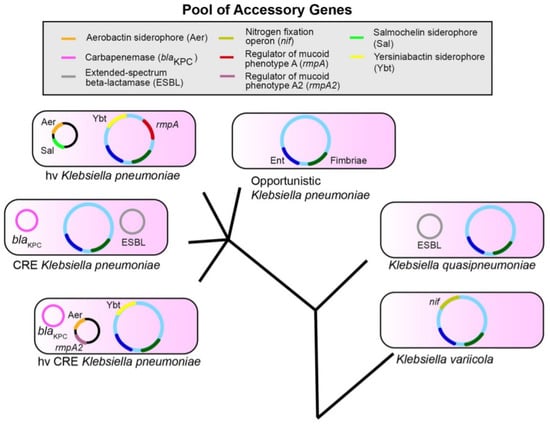
Figure 1.
K. pneumoniae, K. variicola and K. quasipneumoniae are three species that share a set of accessory genes. Ent (Enterobactin, blue) and Fimbriae (dark green) represent conserved genes. The accessory genes are shown as examples, they are not a definitive list. The evolutionary tree is not drawn to scale [1].
These accessory genes can also be combined to form new pathotypes (hvCRE) as they can be shared between species by THG [1,2]. Therefore, the accessory genome reveals the virulence of a bacterium and its study is essential to understand the clinical features of an infectious disease. Moreover, this type of genome can evolve due to the changes in the genome of the pathogen, or by the acquisition of new genes for resistance or variation in its expression. Table 1 shows K. pneumoniae genes that determine the development of antimicrobial resistance with their respective enzymes or target molecules [3]. The high number of genes encoding for β-lactamases supports that they are crucial to the antibiotic resistance.

Table 1.
Antimicrobial resistance genes in the genome of K. pneumoniae [1].
In recent decades, K. pneumoniae has acquired a wide variety of antibiotic resistance mechanisms and, thus, the nosocomial infections it can cause are particularly problematic. This increases the complexity of detecting resistant strains and assigning the appropriate treatment for each bacterium. In a healthcare center, there are several potential foci that facilitate the transmission of K. pneumoniae such as: person-to-person contact between health workers and patients, contaminated surfaces, instrumentation, and some medical techniques (endoscopy). Diseases that weaken the patient, such as cancer, diabetes mellitus, some neurological disorders, or fluid and electrolyte disorders may be classified as risk factors for infection. Despite K. pneumoniae being part of the normal microbiota of the intestine, it is able to colonize other mucosal surfaces, including the nasopharyngeal, urinary, and gastrointestinal tracts, leading to serious infectious diseases in immunocompromised patients [1].
2. Main Resistance Mechanisms in K. pneumoniae
K. pneumoniae has experienced an increase in resistance to different families of antibiotics. Most of these antibiotic resistance mechanisms (AMR) have been acquired through horizontal gene transfer (HGT) conferring high level resistance to antibiotics of the type β-lactams and quinolones.
- (a)
- Mechanisms of antibiotic resistance to β-lactam antibiotics
One of the resistance mechanisms in K. pneumoniae consists in the alteration of penicillin binding proteins (PBPs), enzymes that catalyze peptidoglycan synthesis and specific target of the β-lactam antibiotics. Modifications in their structure decrease the affinity of PBPs for the β-lactam antibiotics, increasing resistance to them [4,5]. Moreover, outer membrane (OM) permeability modifications are critical in K. pneumoniae resistance (hydrophilic drugs). OM is a barrier that must be crossed before it can bind to PBPs [5]. The antibiotic molecules must use porins (OmpK35 and OmpK36), which are reduced in number or modified, decreasing the permeability of the membrane [4,6]. Another common mechanism in K. pneumoniae strains are active ejection pumps like AcrAB and OqxAB pumps [4].
- (b) Quinolone resistance
Mechanisms of fluoroquinolone resistance are due to point mutations in specific areas of DNA gyrase (genes gyrA and gyrB) and topoisomerase IV (genes parC and parE), called quinolone resistance determining region. The first target for quinolone resistance in Gram-negative micro-organisms is gyrA and then parC. Several simultaneous mutations in these genes increase high level resistance [7]. In addition, resistance mechanisms acquired by mobile genetic elements, such as the acquisition of Qnr (quinolone resistance) proteins that protect DNA gyrase and topoisomerase IV. Altered permeability and the presence of efflux pumps are also mechanisms involved in quinolone resistance [8,9].
3. Outbreaks Caused by Resistant Strains of K. pneumoniae
More than 100 different AMR acquired genes have been identified in K. pneumoniae. The presence of SHV-1 penicillinase on the chromosome confers intrinsic resistance to ampicillin, amoxicillin and ticarcillin [2]. Resistance of enterobacteria to the β-lactam antibiotics is associated to the presence of genes encoding for β-lactamases [2,3]. These β-lactamases are frequently encoded by the bla genes and are part of the CTX-M, TEM, SHV and OXA families, sharing different hydrolytic profiles [2]. In the TEM family, mutations of individual nucleotides in genes encoding β-lactamases of the TEM-1 type change the phenotype of BLEE. A wide diversity of members of the TEM family is known, which facilitates the search for transmission of individual resistance genes between strains [2,10]. The amino acid sequence of the SHV-1 family has a high rate of coincidence with TEM-1, sharing its molecular structure. The CTX-M family shows greater activity against cefotaximes than cephtazidime (third-generation cephalosporins), because of the geometry of the antibiotic binding site, which allows efficient recognition of cefotaxime, but not of cephtazidime (a larger molecule). The CTX-M producing strains have the same clinical implications as the production of other BLEE. Nowadays, the number of variants of this family is increasing [2,10]. The main members of the OXA family show low level hydrolytic activity towards clavulanic acid and high level towards carbapenemes. They are the most complex strains to detect and, due to their extensive resistance to β-lactams, therapy has been limited, resulting in high mortality (conditioned by host, type of infection and treatment administered). OXA-48 producing K. pneumoniae is dangerously common in healthcare centers. These are the strains that cause most of the bacteremia outbreaks in hospitals throughout Europe and the Middle East.
The European Commission and the European Centre for Disease Prevention and Control (ECDC) have proposed to increase surveillance and control of outbreaks, in order to reduce the impact of infectious diseases caused by multi-drug resistant organisms on the population [2,10,11]. K. pneumoniae resistant strains to multiple drugs are being more frequently isolated, and this leads to the phenotyping techniques becoming little sensitive or unspecific, making it difficult to identify the strain causing the nosocomial disease. Table 2 shows the main families of plasmids that confer resistance to clinically relevant antibiotics, such as extended spectrum cephalosporins, fluoroquinolones and aminoglycosides. The number of references obtained for TEM, KPC and CTX-M type enzymes is very high compared to the rest of the enzymes, supporting their importance for the health field and the scientific community. In the 1970s, most outbreaks of healthcare associated infections (HAI) caused by K. pneumoniae were due to aminoglycoside resistant strains. Between 1990 and 2000, outbreaks caused by BLEE producing strains were described, while those caused by carbapenemic resistant strains were described from 2000 onwards. Today, strains producing carbapenemics such as KPC and NDM-1, are significant in clinical practice [11].

Table 2.
Review of resistant plasmid families associated with AmpC, BLEE, 16S rRNA methylases, Qnr proteins and MBLs in K. pneumoniae.
On the one hand, K. pneumoniae NDM-1 is a zinc dependent metalo-β-lactamase that was first identified in 2009 from a New Delhi patient. This enzyme, anchored to the external membrane, can capture the zinc present in the host. In addition, vesicles can carry the active enzyme NDM-1 by exocytosis. Therefore, it can reach other bacteria and bind to the membrane acting as if it were also expressed by other bacteria, making them temporarily resistant [12]. Furthermore, NDM-1 is encoded by the plasmid gene blaNMD-1 and can reach other strains through HGT. Strains producing this enzyme hydrolyze all β-lactam antibiotics, except aztreonam. Among the possible inhibitors studied, hesperidins show high activity against NDM-1. Due to the low toxicity hesperidins contain, they are on the list of possible NDM-1 inhibitor drugs [12]. Since 2009, nearly 650 articles have been reported in PubMed on the presence of NDM-1 in Klebsiella.
On the other hand, KPC is an enzyme capable of conferring resistance to carbapenemics, as well as inactivating penicillins, cephalosporins or monobactamics. It was first identified in 2001 in the United States from a superbug that had suffered a mutation that conferred resistance to multiple drugs [13]. Bacteria with KPC enzymes are more virulent because they have acquired resistance to multiple antibiotics, leading to a high mortality rate in immunocompromised patients. Research confirms that previous use of carbapenemics increases the likelihood of a patient developing a HAI caused by KPC-producing bacteria [13]. However, there is some controversy in the scientific community about this claim, as a study on the clinical impact of carbapenemically resistant K. pneumoniae concluded that previous use of fluoroquinolone and broad-spectrum cephalosporin was independent of infections with KPC producing organisms [14].
4. Conclusions
The massive and uncontrolled use of antimicrobials in clinical practice (mainly the β-lactam type) has contributed to the accelerated development of antimicrobial resistance. The increase in nosocomial infections caused by BLEE-producing enterobacteria such as K. pneumoniae is generating a global economic stress due to the numerous treatments that have been compromised. Therefore, the main objective is not to expand the therapeutic arsenal, but to develop other techniques far from the direct use of antimicrobials, reducing the effects and impact of multi-drug resistant organisms on the population.
Author Contributions
Conceptualization S.M., formal analysis, C.M., writing C.M., S.M. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ESBL | Extended Spectrum β-Lactamase |
References
- Martin, R.; Bachman, M. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.; Holt, K. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.; Holt, K. Klebsiella pneumoniae Population Genomics and Antimicrobial-Resistant Clones. Trends Microbiol. 2016, 24, 944–956. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Kaczanowska, M.; Rydén-Aulin, M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007, 71, 477–494. [Google Scholar] [CrossRef] [PubMed]
- Villa, L.; Feudi, C.; Fortini, D.; Garcia-Fernandez, A.; Carattoli, A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob. Agents Chemother. 2014, 58, 1707–1712. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Kawamura, Y.; Ezaki, T.; Balakrish Nair, G.; Iida, K.I.; Yoshida, S.I. Alteration in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant Shigella dysenteriae serotype 1 clinical isolates from Kolkata, India. Antimicrob. Agents Chemother. 2005, 49, 1660–1661. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Martinez, L.; Hernandez-Allas, S.; Alberti, S.; Tomas, J.; Benedi, V.; Jacoby, G. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 1996, 40, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Pons, M.; Gomes, C. Transferable mechanisms of quinolone resistance. Int. J. Antimicrob. Agents 2012, 40, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Gao, J.; Tang, W. Nosocomial infection and its molecular mechanisms of antibiotic resistance. Biosci. Trends 2016, 10, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.; Harris, P.; Henderson, A.; Paterson, D. Treatment of infections by OXA-48-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Chen, J.; Xiao, B.; Kang, X.; Lao, X.; Zheng, H. Discovery of NDM-1 inhibitors from natural products. J. Glob. Antimicrob. Resist. 2019, 18, 80–87. [Google Scholar] [CrossRef]
- Weisenberg, S.; Morgan, D.; Espinal-Witter, R.; Larone, D. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn. Microbiol. Infect. Dis. 2009, 64, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Gasink, L.; Edelstein, P.; Lautenbach, E.; Synnestvedt, M.; Fishman, N. Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Infect. Control Hosp. Epidemiol. 2009, 30, 1180–1185. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).