Effect of Common Bean Consumption on the Gut Associated Microbiome in an In Vivo Screening Model for Breast Cancer †
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Genotyping
2.2. Screening Assay
2.3. Histological Assessment
2.4. Western-Blot-Based Immuno-Nanocapillary Electrophoresis
2.5. Microbiota Characterization
2.6. Statistical Analyses
3. Results and Discussion
3.1. Development of a Mammary Cancer Screening Model
3.2. Effect of Common Bean on the Expansion of Mammary Pathologies In Vivo
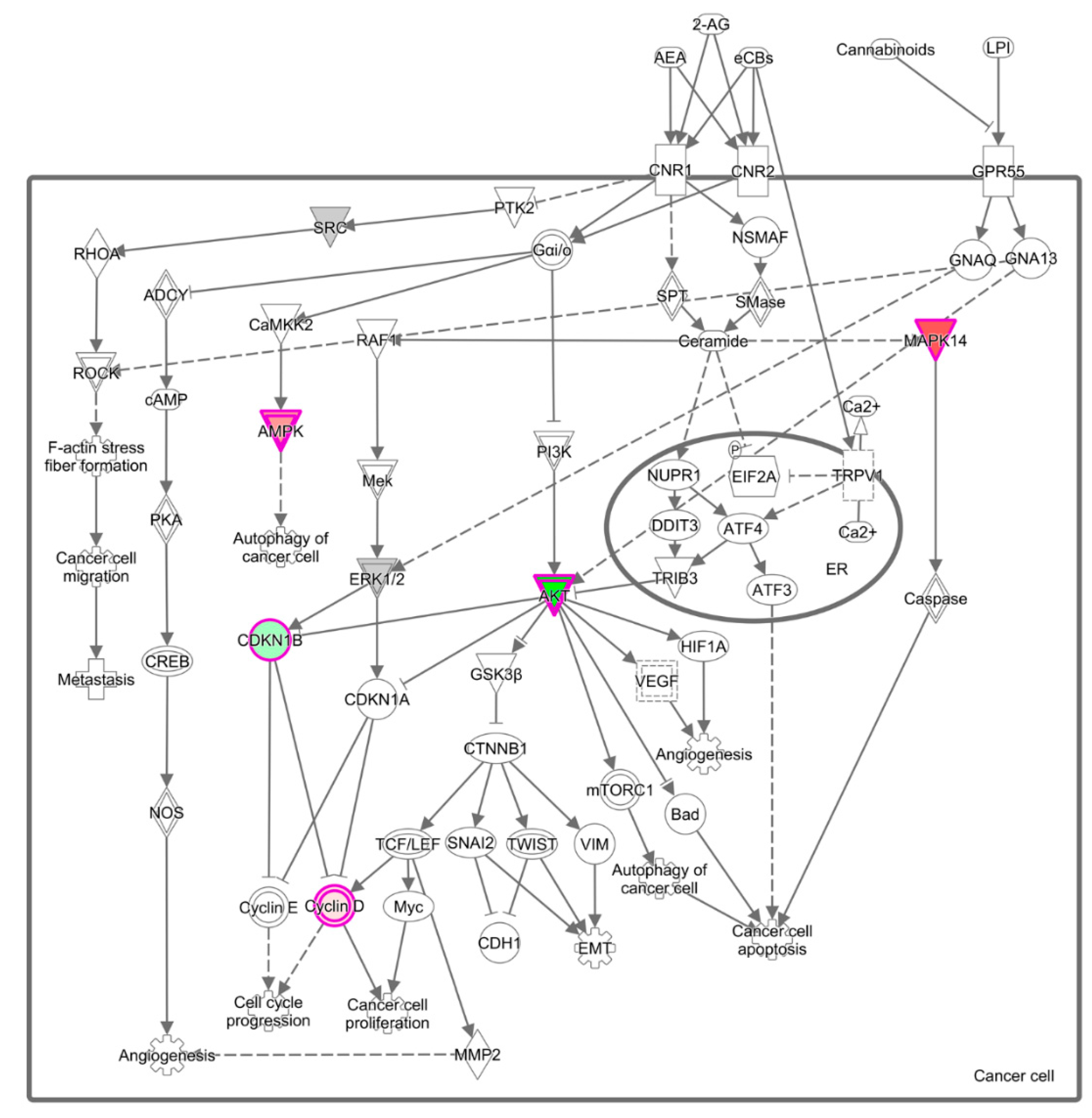
3.3. Effect of Common Bean on Cell Signaling Pathways in Mammary Pathologies In Vivo
3.4. Effect of Common Bean on the Gut Associated Microbiome
4. Concluding Comments
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- NIH. The Science Behind Healthy Eating Patterns; National Institutes of Health: Bethesda, MD, USA, 2020. [Google Scholar]
- 2020–2030 Strategic Plan for NIH Nutrition Research. 2020. Available online: https://www.niddk.nih.gov/about-niddk/strategic-plans-reports/strategic-plan-nih-nutrition-research (accessed on 15 October 2020).
- Frame, L.A.; Costa, E.; Jackson, S.A. Current Explorations of Nutrition and the Gut Microbiome: A Comprehensive Evaluation of the Review Literature. Nutr. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- McBurney, M.I.; Davis, C.; Fraser, C.M.; Schneeman, B.O.; Huttenhower, C.; Verbeke, K.; Walter, J.; Latulippe, M.E. Establishing What Constitutes a Healthy Human Gut Microbiome: State of the Science, Regulatory Considerations, and Future Directions. J. Nutr. 2019, 149, 1882–1895. [Google Scholar] [CrossRef] [PubMed]
- Bilotta, A.J.; Cong, Y. Gut microbiota metabolite regulation of host defenses at mucosal surfaces: implication in precision medicine. Precis. Clin. Med. 2019, 2, 110–119. [Google Scholar] [CrossRef]
- Rodgers, G.P.; Collins, F.S. Precision Nutrition-the Answer to “What to Eat to Stay Healthy”. JAMA 2020. [Google Scholar] [CrossRef]
- Adebamowo, C.A.; Cho, E.; Sampson, L.; Katan, M.B.; Spiegelman, D.; Willett, W.C.; Holmes, M.D. Dietary flavonols and flavonol-rich foods intake and the risk of breast cancer. Int. J. Cancer 2005, 114, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Murtaugh, M.A.; Sweeney, C.; Giuliano, A.R.; Herrick, J.S.; Hines, L.; Byers, T.; Baumgartner, K.B.; Slattery, M.L. Diet patterns and breast cancer risk in Hispanic and non-Hispanic white women: the Four-Corners Breast Cancer Study. Am. J. Clin. Nutr. 2008, 87, 978–984. [Google Scholar] [CrossRef]
- Thompson, M.D.; Mensack, M.M.; Jiang, W.; Zhu, Z.; Lewis, M.R.; McGinley, J.N.; Brick, M.A.; Thompson, H.J. Cell signaling pathways associated with a reduction in mammary cancer burden by dietary common bean (Phaseolus vulgaris L.). Carcinogenesis 2012, 33, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.D.; Thompson, H.J.; Brick, M.A.; McGinley, J.N.; Jiang, W.; Zhu, Z.; Wolfe, P. Mechanisms associated with dose-dependent inhibition of rat mammary carcinogenesis by dry bean (Phaseolus vulgaris, L.). J. Nutr. 2008, 138, 2091–2097. [Google Scholar] [CrossRef]
- Mensack, M.M.; Fitzgerald, V.K.; Lewis, M.R.; Thompson, H.J. Characterization of low molecular weight chemical fractions of dry bean (Phaseolus vulgaris) for bioactivity using Caenorhabditis elegans longevity and metabolite fingerprinting. J. Agric. Food Chem. 2010, 58, 6697–6705. [Google Scholar] [CrossRef][Green Version]
- Davie, S.A.; Maglione, J.E.; Manner, C.K.; Young, D.; Cardiff, R.D.; MacLeod, C.L.; Ellies, L.G. Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res. 2007, 16, 193–201. [Google Scholar] [CrossRef]
- Guy, C.T.; Cardiff, R.D.; Muller, W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell Biol. 1992, 12, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Neil, E.S.; McGinley, J.N.; Fitzgerald, V.K.; Lauck, C.A.; Tabke, J.A.; Streeter-McDonald, M.R.; Yao, L.; Broeckling, C.D.; Weir, T.L.; Foster, M.T.; et al. White Kidney Bean (Phaseolus Vulgaris L.) Consumption Reduces Fat Accumulation in a Polygenic Mouse Model of Obesity. Nutrients 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- McGinley, J.N.; Thompson, H.J. Quantitative assessment of mammary gland density in rodents using digital image analysis. Biol. Proced. Online 2011, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; McGinley, J.N.; Thompson, H.J. A comparison of the histopathology of premalignant and malignant mammary gland lesions induced in sexually immature rats with those occurring in the human. Lab. Invest. 2000, 80, 221–231. [Google Scholar] [CrossRef]
- Thompson, H.J.; McGinley, J.N.; Neil, E.S.; Brick, M.A. Beneficial Effects of Common Bean on Adiposity and Lipid Metabolism. Nutrients 2017, 9, 998. [Google Scholar] [CrossRef]
- Lee, D.M.; Battson, M.L.; Jarrell, D.K.; Hou, S.; Ecton, K.E.; Weir, T.L.; Gentile, C.L. SGLT2 inhibition via dapagliflozin improves generalized vascular dysfunction and alters the gut microbiota in type 2 diabetic mice. Cardiovasc. Diabetol. 2018, 17, 62. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 2019. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Lin, E.Y.; Pollard, J.W. Macrophages: modulators of breast cancer progression. Novartis Found Symp. 2004, 256, 158–168. [Google Scholar]
- Kovalchuk, O.; Kovalchuk, I. Cannabinoids as anticancer therapeutic agents. Cell Cycle 2020, 19, 961–989. [Google Scholar] [CrossRef]
- Takeda, S.; Okajima, S.; Miyoshi, H.; Yoshida, K.; Okamoto, Y.; Okada, T.; Amamoto, T.; Watanabe, K.; Omiecinski, C.J.; Aramaki, H. Cannabidiolic acid, a major cannabinoid in fiber-type cannabis, is an inhibitor of MDA-MB-231 breast cancer cell migration. Toxicol. Lett. 2012, 214, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Hermanson, D.J.; Marnett, L.J. Cannabinoids, endocannabinoids, and cancer. Cancer Metastasis Rev. 2011, 30, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.R.; McGee, R.; Vandemark, G.; Brick, M.; Thompson, H.J. Dietary Fiber Analysis of Four Pulses Using AOAC 2011.25: Implications for Human Health. Nutrients 2016, 8, 829. [Google Scholar] [CrossRef] [PubMed]
- Thompson, H.J.; Brick, M.A. Perspective: Closing the Dietary Fiber Gap: An Ancient Solution for a 21st Century Problem. Adv. Nutr. 2016, 7, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Kleintop, A.E.; Echeverria, D.; Brick, L.A.; Thompson, H.J.; Brick, M.A. Adaptation of the AOAC 2011.25 integrated total dietary fiber assay to determine the dietary fiber and oligosaccharide content of dry edible beans. J. Agric. Food Chem. 2013, 61, 9719–9726. [Google Scholar] [CrossRef] [PubMed]
- Muccioli, G.G.; Naslain, D.; Bäckhed, F.; Reigstad, C.S.; Lambert, D.M.; Delzenne, N.M.; Cani, P.D. The endocannabinoid system links gut microbiota to adipogenesis. Mol. Syst. Biol. 2010, 6, 392. [Google Scholar] [CrossRef]
- Manca, C.; Boubertakh, B.; Leblanc, N.; Deschenes, T.; Lacroix, S.; Martin, C.; Houde, A.; Veilleux, A.; Flamand, N.; Muccioli, G.G.; et al. Germ-free mice exhibit profound gut microbiota-dependent alterations of intestinal endocannabinoidome signaling. J. Lipid Res. 2020, 61, 70–85. [Google Scholar] [CrossRef]
- Lu, H.-C.; Mackie, K. An Introduction to the Endogenous Cannabinoid System. Biol. Psychiatry 2016, 79, 516–525. [Google Scholar] [CrossRef]
- McGinley, J.N.; Fitzgerald, V.K.; Neil, E.S.; Omerigic, H.M.; Heuberger, A.L.; Weir, T.L.; McGee, R.; Vandemark, G.; Thompson, H.J. Pulse Crop Effects on Gut Microbial Populations, Intestinal Function, and Adiposity in a Mouse Model of Diet-Induced Obesity. Nutrients 2020, 12, 593. [Google Scholar] [CrossRef]
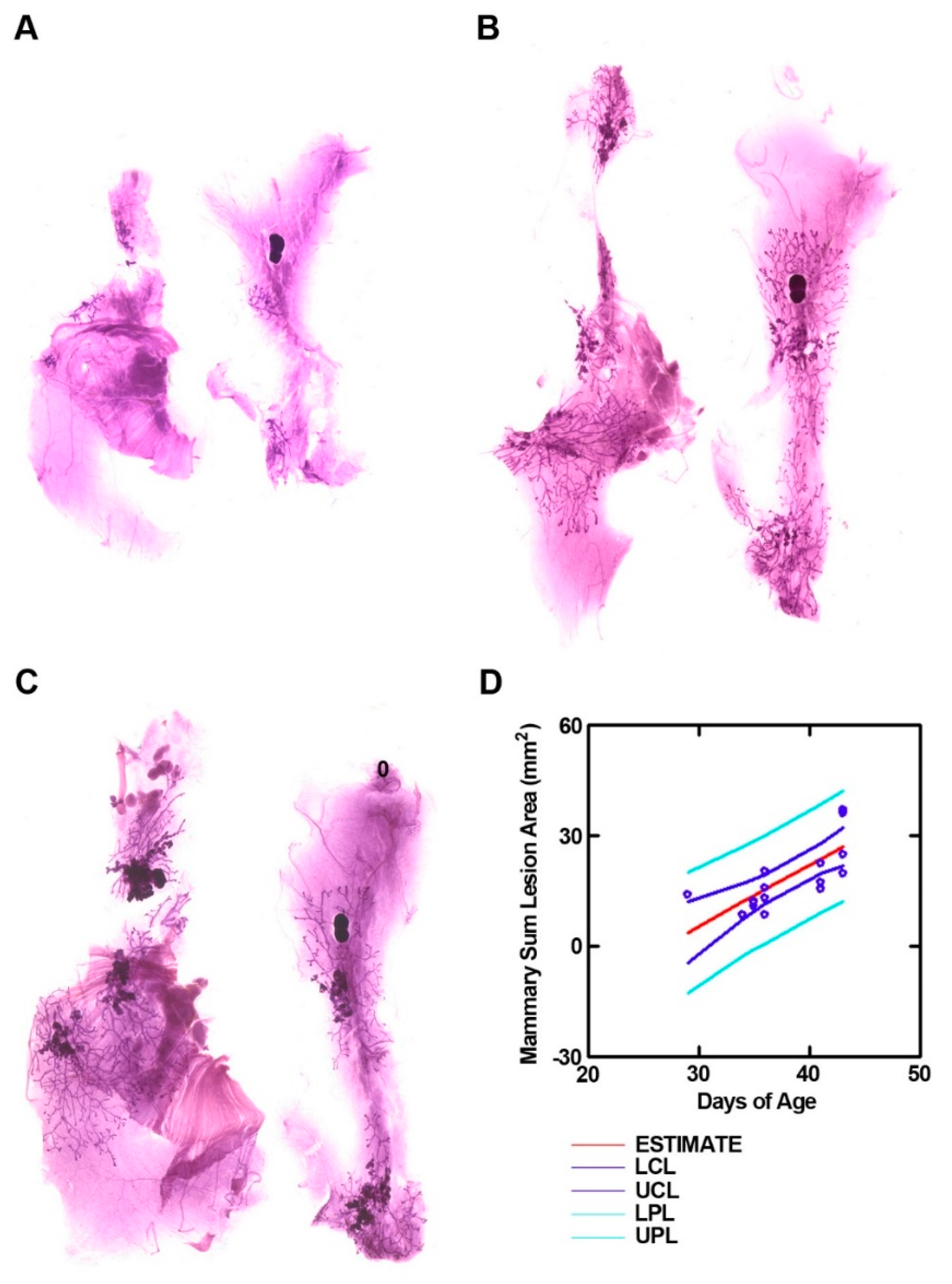
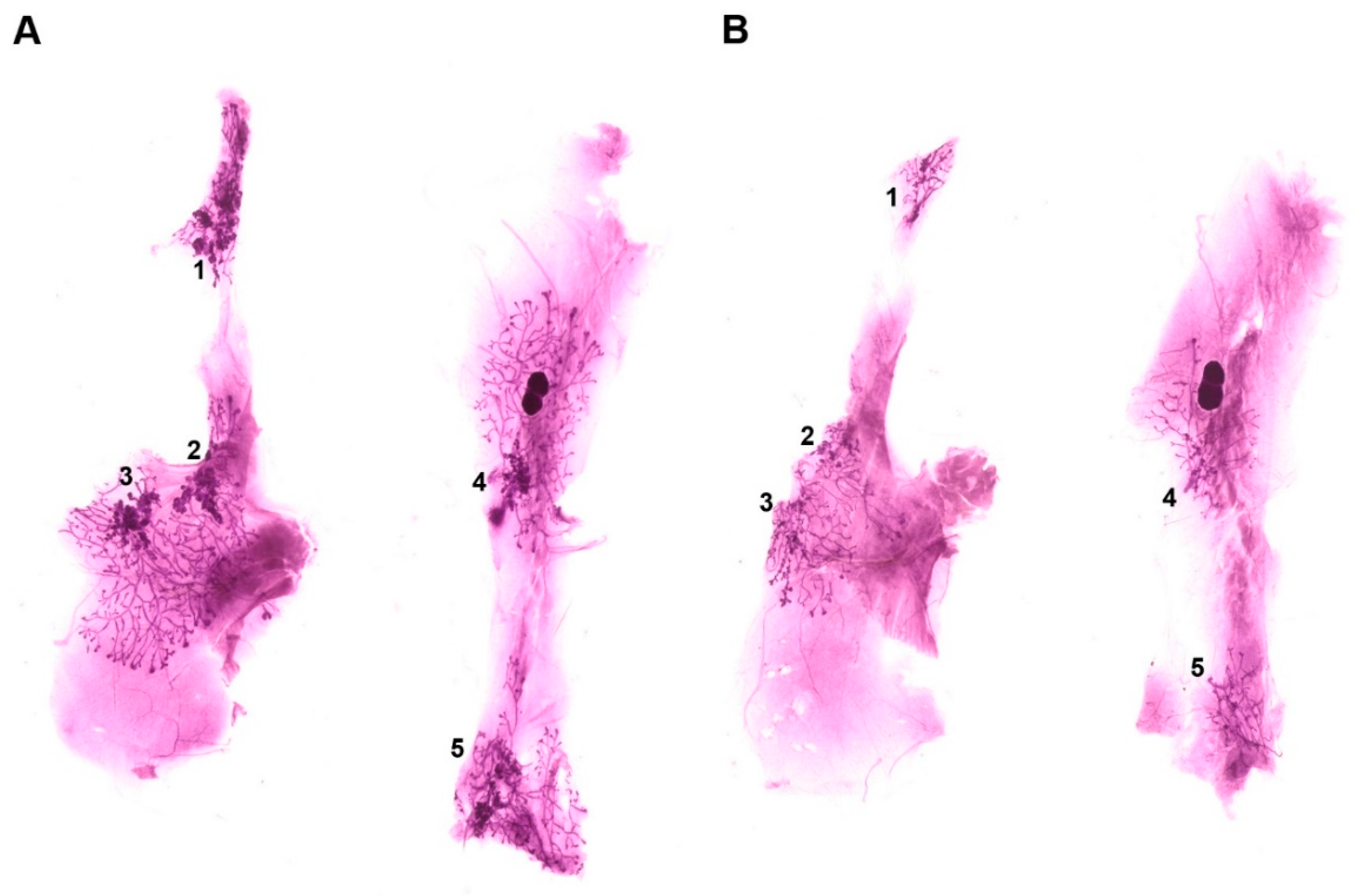
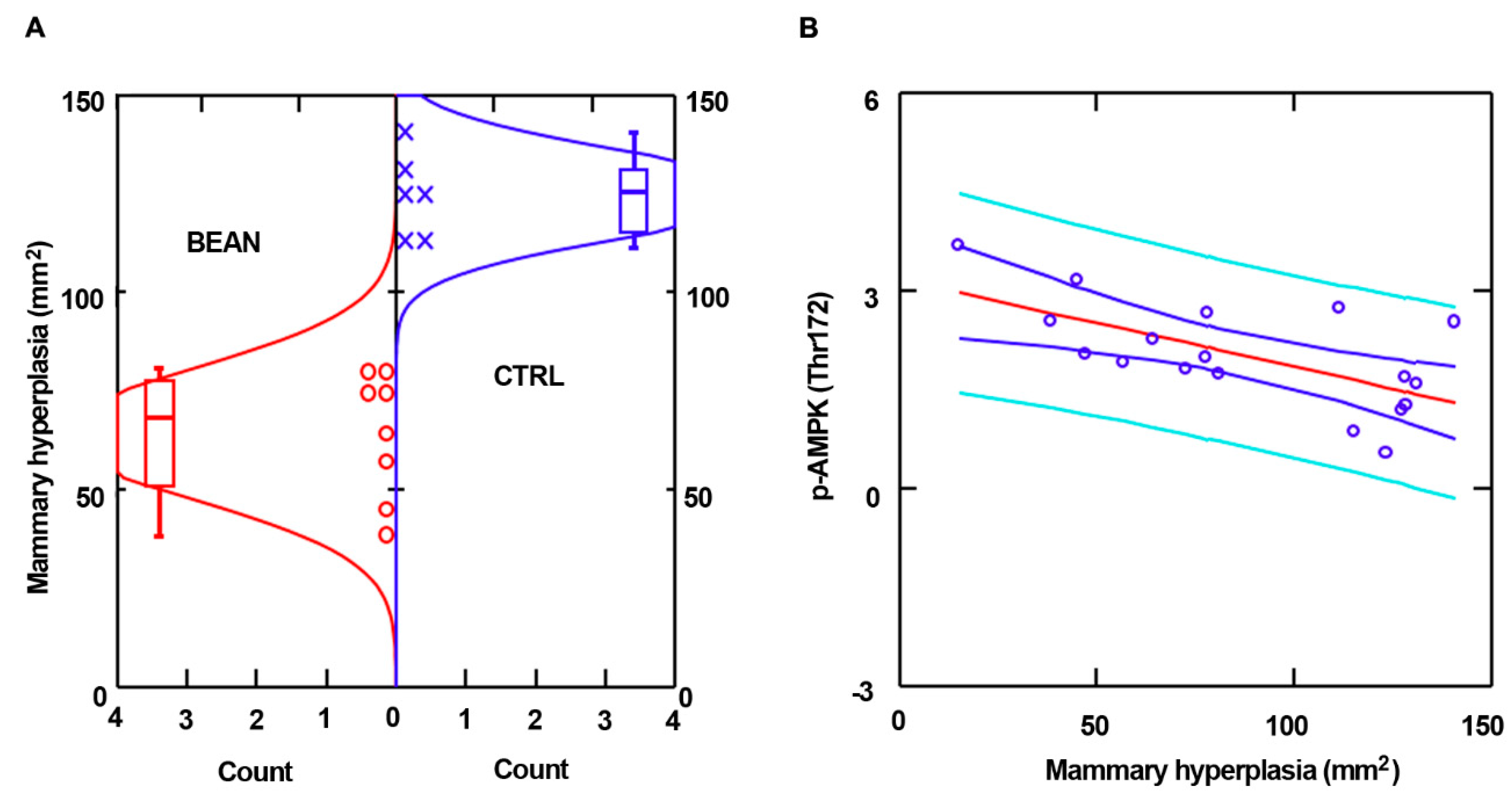
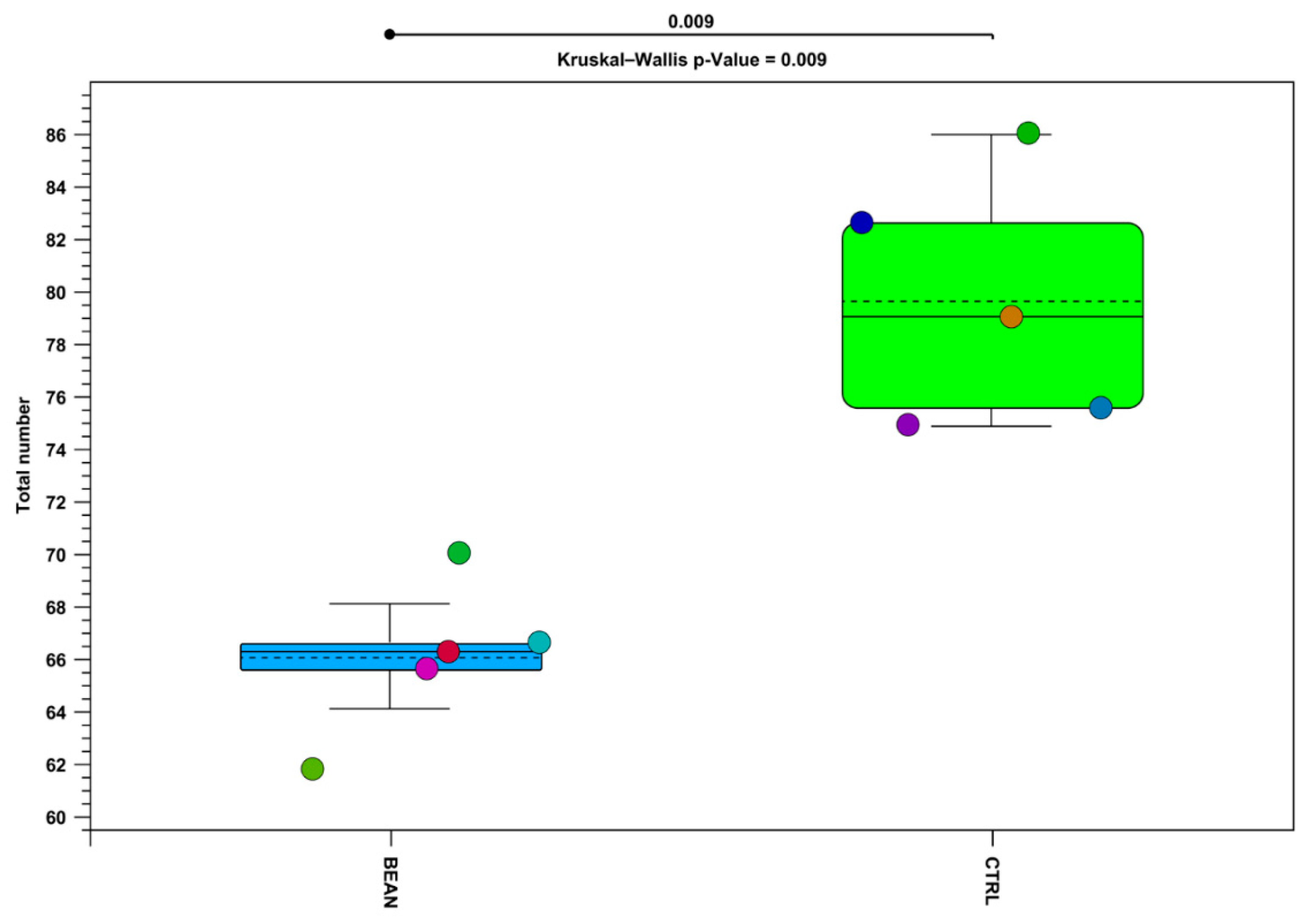
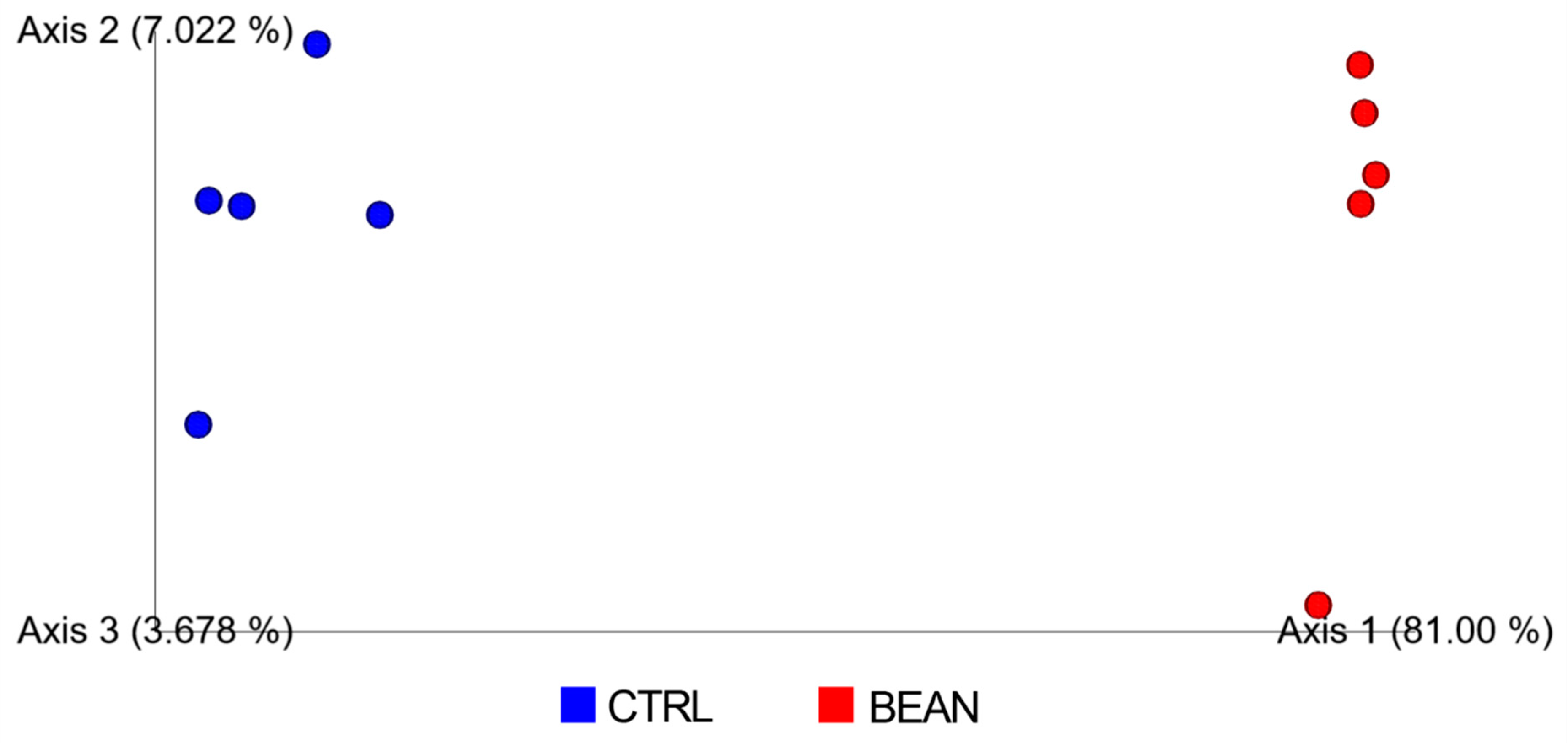
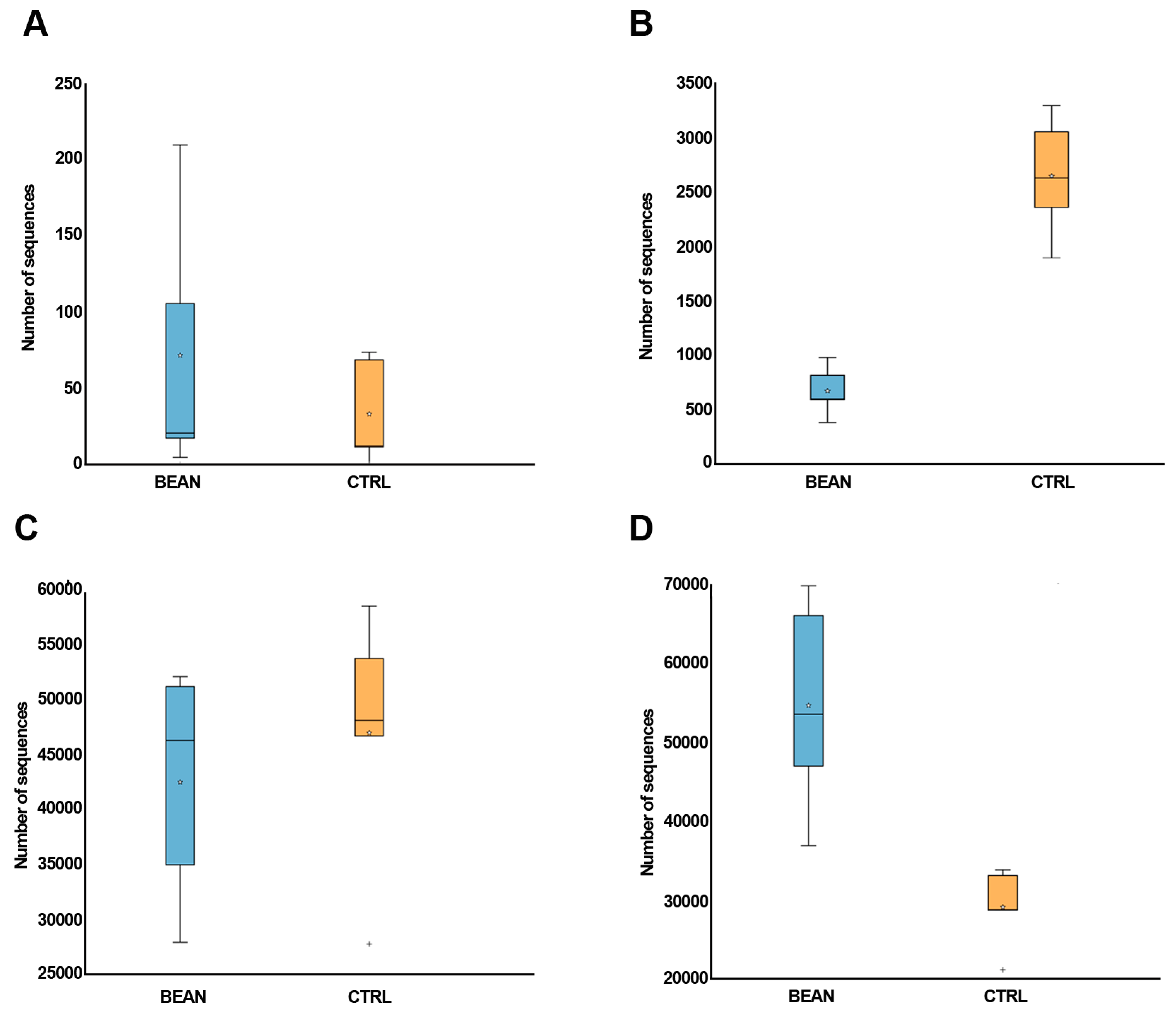
| Sum Lesion Area (mm2) | CTRL (n = 9) | Bean (n = 10) | p-Value |
|---|---|---|---|
| Mammary gland 1 | 7.64 ± 1.00 | 4.00 ± 0.66 | 0.007 |
| Mammary gland 2 | 3.37 ± 0.74 | 1.80 ± 0.54 | 0.092 |
| Mammary gland 3 | 5.38 ± 1.10 | 2.26 ± 0.45 | 0.014 |
| Mammary gland 4 | 6.00 ± 2.06 | 1.66 ± 0.33 | 0.043 |
| Mammary gland 5 | 5.86 ± 1.79 | 1.95 ± 0.29 | 0.028 |
| Phylum | log2FC | p_Value | FDR | Ctrl_Log2_Mean | Bean_Log2_Mean |
|---|---|---|---|---|---|
| (Unknown Phylum) Bacteria | −0.051354766 | 0.038652403 | 0.064420671 | −6.581462293 | −6.632817059 |
| Actinobacteria | −0.142413323 | 0.016577252 | 0.033154504 | −6.501442867 | −6.64385619 |
| Bacteria | −0.094320142 | 0.064927787 | 0.081159733 | −6.483667922 | −6.577988064 |
| Bacteroidetes | 1.784902055 | 0.000195928 | 0.000979641 | −2.165674619 | −0.380772565 |
| Candidatus Saccharibacteria | −0.004295945 | 0.373900966 | 0.373900966 | −6.639560244 | −6.64385619 |
| Deferribacteres | −1.418705665 | 0.011339537 | 0.028348842 | −5.066253673 | −6.484959337 |
| Firmicutes | −1.759348332 | 0.000525378 | 0.001751259 | −0.536789118 | −2.296137451 |
| Proteobacteria | −1.94107296 | 5.69E−05 | 0.000568878 | −3.558157983 | −5.499230943 |
| Tenericutes | −0.007939984 | 0.373900966 | 0.373900966 | −6.635916206 | −6.64385619 |
| Verrucomicrobia | 1.403597718 | 0.059447941 | 0.081159733 | −6.64385619 | −5.240258472 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, H.J.; McGinley, J.N.; Neil, E.S.; Weir, T.L. Effect of Common Bean Consumption on the Gut Associated Microbiome in an In Vivo Screening Model for Breast Cancer. Proceedings 2020, 61, 14. https://doi.org/10.3390/IECN2020-07008
Thompson HJ, McGinley JN, Neil ES, Weir TL. Effect of Common Bean Consumption on the Gut Associated Microbiome in an In Vivo Screening Model for Breast Cancer. Proceedings. 2020; 61(1):14. https://doi.org/10.3390/IECN2020-07008
Chicago/Turabian StyleThompson, Henry J., John N. McGinley, Elizabeth S. Neil, and Tiffany L. Weir. 2020. "Effect of Common Bean Consumption on the Gut Associated Microbiome in an In Vivo Screening Model for Breast Cancer" Proceedings 61, no. 1: 14. https://doi.org/10.3390/IECN2020-07008
APA StyleThompson, H. J., McGinley, J. N., Neil, E. S., & Weir, T. L. (2020). Effect of Common Bean Consumption on the Gut Associated Microbiome in an In Vivo Screening Model for Breast Cancer. Proceedings, 61(1), 14. https://doi.org/10.3390/IECN2020-07008






