Peptide-Based Biosensor for Express Diagnostics of Coronavirus Respiratory Infections †
Abstract
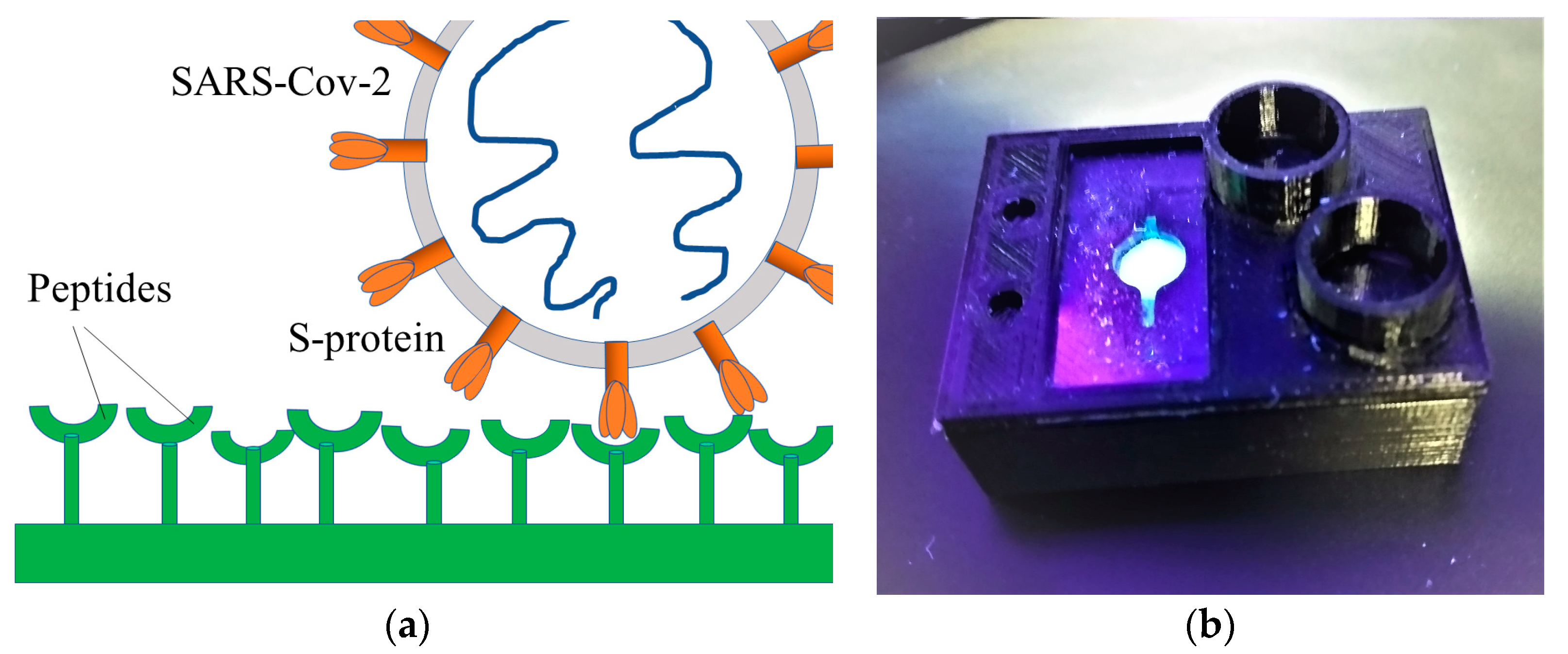
:1. Introduction
2. Materials and Methods
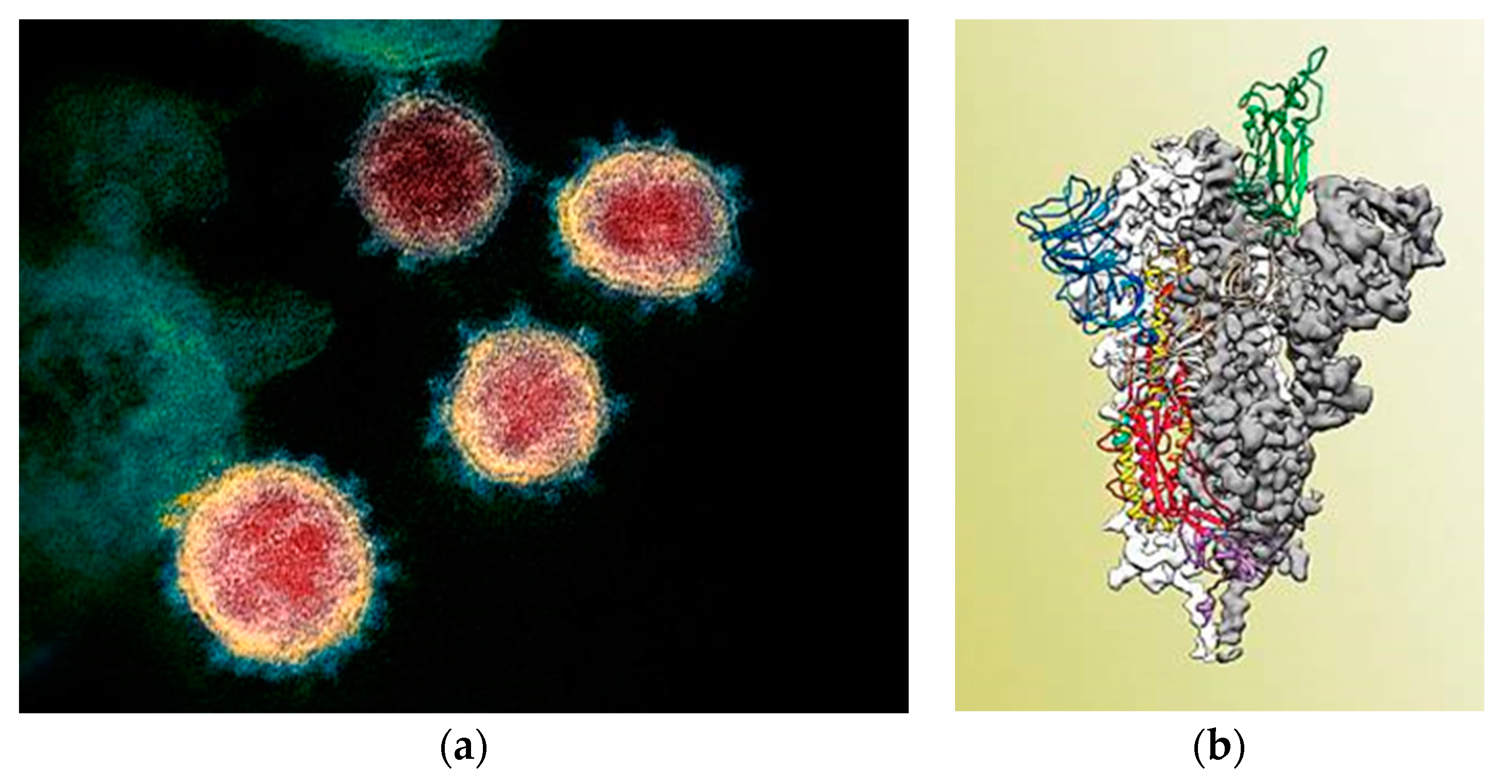
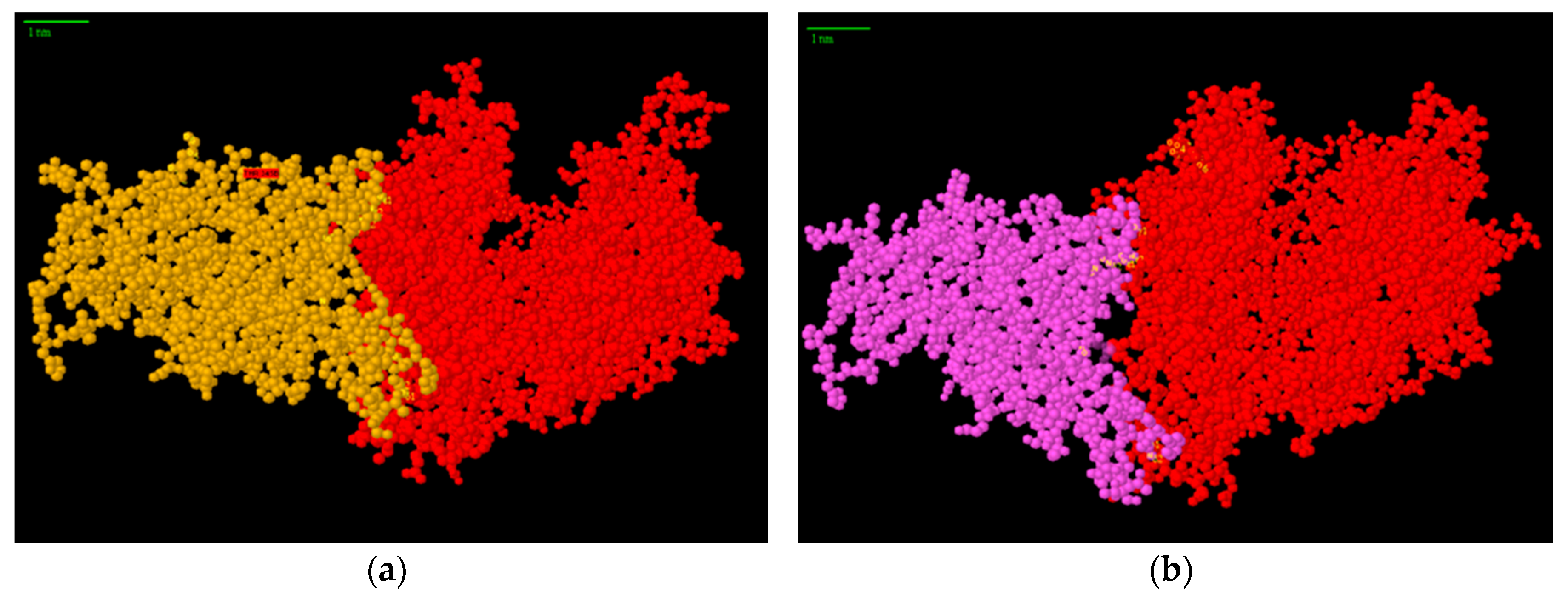
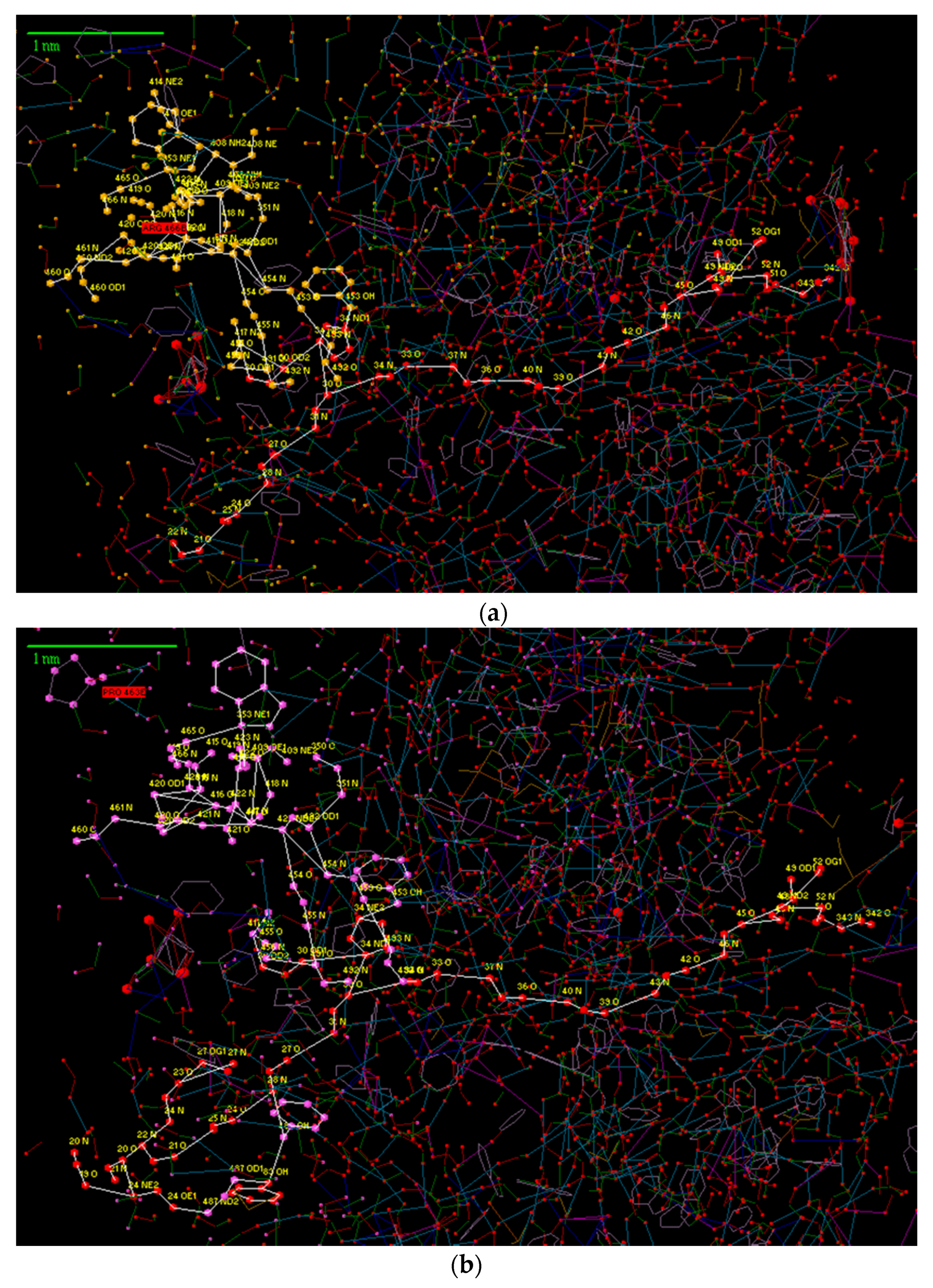
3. Results and Discussion
References
- WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 25 October 2020).
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Available online: http://protein-3d.ru (accessed on 16 April 2020).
- Karasev, V.A.; Luchinin, V.V. Introduction into Design of Bionic Nanosystems; Physmatlit: Moscow, Russia, 2011; 464p. [Google Scholar]
- Protein Data Bank. Available online: https://www.rcsb.org/ (accessed on 15 April 2020).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zimina, T.; Karasev, V.; Luchinin, V.; Kolobov, A.; Mandrik, I.; Sitkov, N.; Kraeva, L.; Shevchenko, N.; Orekhov, Y. Peptide-Based Biosensor for Express Diagnostics of Coronavirus Respiratory Infections. Proceedings 2020, 60, 52. https://doi.org/10.3390/IECB2020-07059
Zimina T, Karasev V, Luchinin V, Kolobov A, Mandrik I, Sitkov N, Kraeva L, Shevchenko N, Orekhov Y. Peptide-Based Biosensor for Express Diagnostics of Coronavirus Respiratory Infections. Proceedings. 2020; 60(1):52. https://doi.org/10.3390/IECB2020-07059
Chicago/Turabian StyleZimina, Tatiana, Vladimir Karasev, Viktor Luchinin, Alexander Kolobov, Ivan Mandrik, Nikita Sitkov, Liudmila Kraeva, Natalia Shevchenko, and Yuri Orekhov. 2020. "Peptide-Based Biosensor for Express Diagnostics of Coronavirus Respiratory Infections" Proceedings 60, no. 1: 52. https://doi.org/10.3390/IECB2020-07059
APA StyleZimina, T., Karasev, V., Luchinin, V., Kolobov, A., Mandrik, I., Sitkov, N., Kraeva, L., Shevchenko, N., & Orekhov, Y. (2020). Peptide-Based Biosensor for Express Diagnostics of Coronavirus Respiratory Infections. Proceedings, 60(1), 52. https://doi.org/10.3390/IECB2020-07059





