1. Introduction
PDMS is one of the most actively developed polymers for microfluidics designing. The fabrication of channel systems in PDMS-poly(dimethylsiloxane) is facilitated by the fact that it can be cast using a suitable mold with sub-0.1 µm fidelity. Being more than just a structural material it also allows the fabrication of devices with useful functionality due to its chemical and physical properties: elasticity, optical transparency, low water permeability, low electrical conductivity, and the possibility of its patterning by lithographic techniques [
1].
Usually, the pathogen’s detection is based on immobilization by using specific antigens, antibodies, labelling molecules, etc. Moreover, several approaches of capturing/concentration/filtration are used for having the significant biomass required for accurate detection. For instance, the on-line SERS detection and accurate identification of suspended bacteria with a detection capability down to a single bacterium has been realized by the nanoaggregate-embedded beads (NAEBs)-dielectrophoresis (DEP)-Raman spectroscopy biosensing strategy [
2].
In a study from 2013, Chrimes et al. [
3] reports the fabrication of microfluidic channels with a size of 150 × 70 μm
2 and a length of 12 mm, made of PDMS on quartz substrate, the latter being chosen due to the very weak Raman signal and interference with the SERS signal from the analyte (yeast
Saccharomyces cerevisiae). The microfluidic system was connected to an injection pump to facilitate the transport of the yeasts through the microfluidic channels. To perform the measurement, the yeasts coated with Ag nanoparticles (AgNPs) were manipulated and immobilized inside the microchannel by using DEP force field. Another study [
4] reports the development of a microfluidic platform for the detection of several pathogenic bacteria using AgNPs as a SERS substrate. In this case, the microfluidic channels were also made in PDMS on glass substrate by means of photolithography method.
We herein propose a practical detection approach as an alternative to conventional molecular biology methods, by developing an experimental setup based on the principle of in situ preparation of silver SERS-active substrate, in a Plexiglas microfluidic detection cell, by using the portable Raman spectrometer as detection tool.
2. Materials and Methods
For bacterial cells activation, aliquot with E. coli TOP10, P. aeruginosa, S. aureus, and E. faecalis, stored at −80 °C for long-term storage, was previously partially thawed at room temperature. After thawing, 1 µL of bacteria was transferred using a sterile plastic loop into a new tube containing 5 mL of liquid Luria Broth (LB) media. The LB medium was previously sterilized by autoclaving at 121 °C for 5 min. The tube containing media and the inoculum was vortexed for 5 s (maximum speed) and then incubated overnight at 37 °C. To obtain pure colonies, 10 µL from overnight culture was then subjected to serial dilution technique. After inoculation of LB medium with the desired dilution, the Petri dishes were incubated 15 min at room temperature and after were transferred to the incubator at 37 °C overnight (at least 20 h).
Standard SERS samples preparation. After incubation, we inspected the Petri dishes and a single colony of bacteria was transferred into 5 mL of sterile LB Broth and grown overnight at 37 °C. After growing, 4 mL of E. faecalis culture were centrifuged at 3000× g for 2 min, in two steps (2 mL of bacterial culture for each step), the supernatant was discarded and the remained pellet was resuspended (washed) in 2 mL of sterile saline solution and gently vortexed. This washing protocol was repeated three times for a thorough discard of the growth components. The working culture was obtained through resuspension of the obtained pellet (after three steps of washing) in 200 µL of sterile saline solution. The working culture was stored at 4 °C until further analysis. If the bacterial suspension was too concentrated, further dilutions were carried out with the same saline solution.
SERS spectra acquisition. The Raman system used to perform the SERS experiments consisted of a BW-TEK portable i-Raman spectrometer (B&W Tek Inc., Lübeck, Germany) equipped with a 532 nm laser line and a total power source of 50 mW, connected through an optical fiber to a BW-TEK optical microscope equipped with 20× objective. A syringe with 3 mL of the bacterial biomass was attached to the two-channel injection pump (NE-4000 Programmable 2 Channel Syringe Pump, NewEra Pump Systems Inc., New York, USA) used in the microfluidic SERS-based detection setup.
3. Results
3.1. Experimental Setup
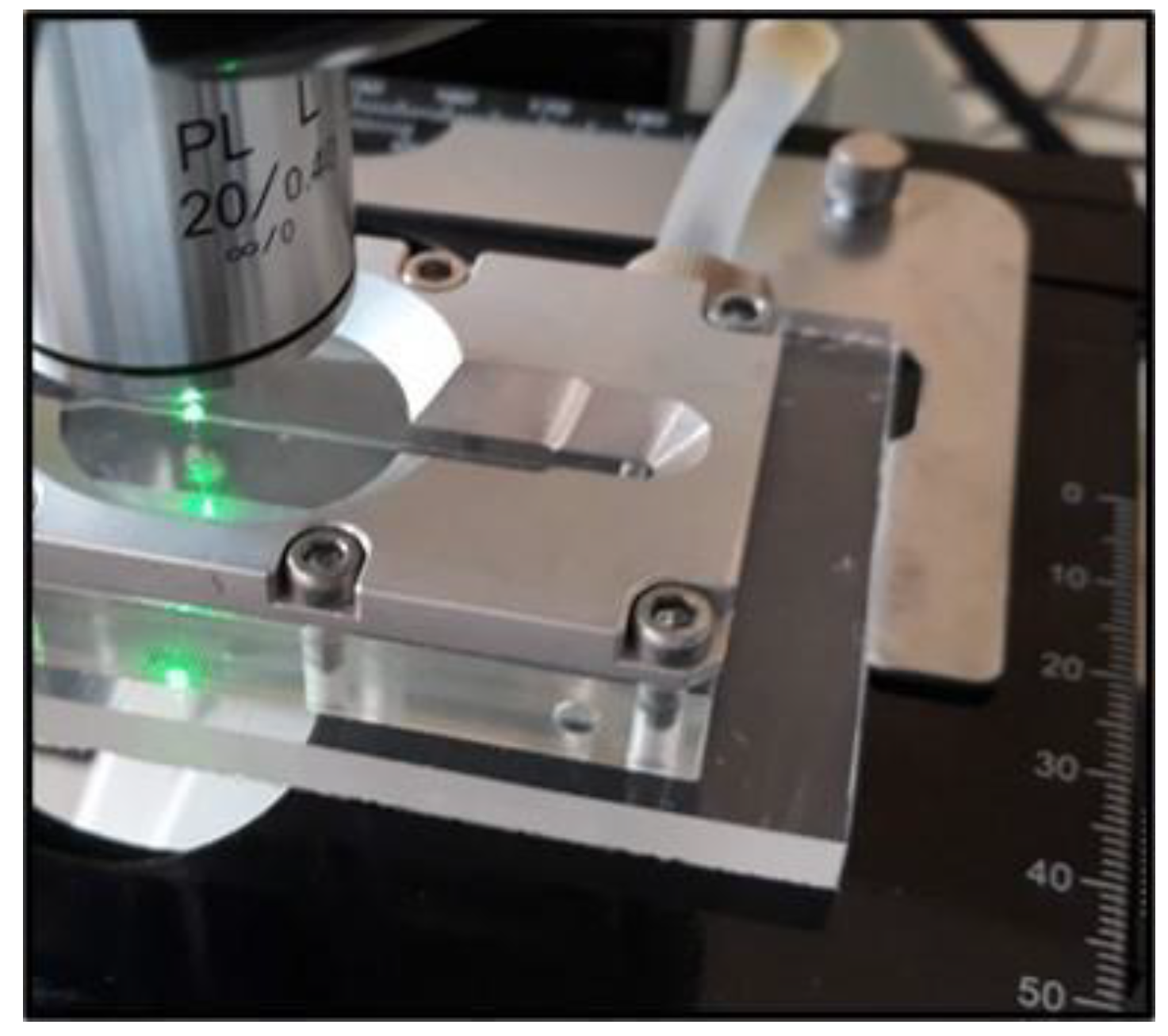
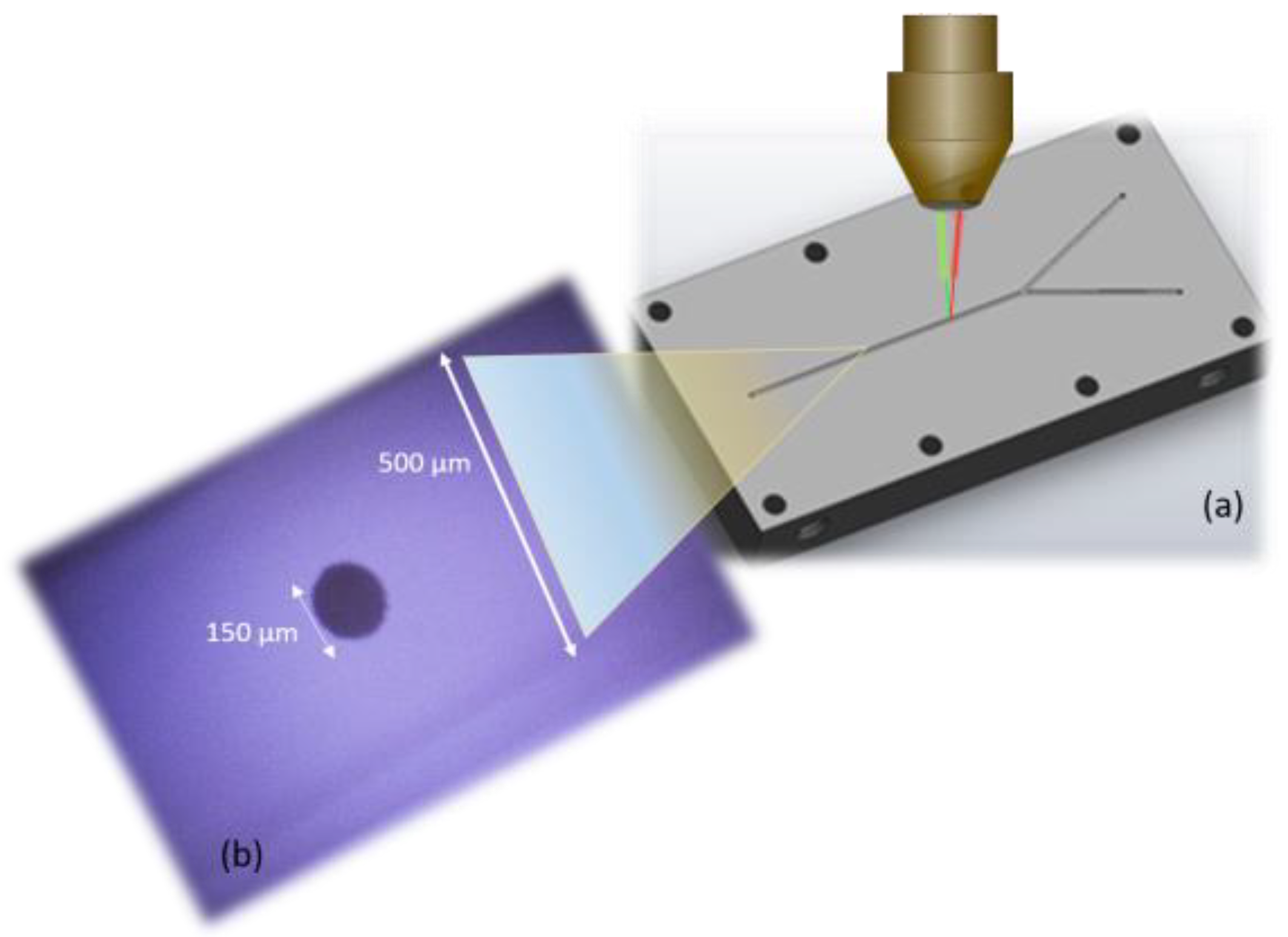
Figure 1 shows the microfluidic cell placed on the table of a Raman microscope, which is connected to both the portable Raman spectrometer equipped with the laser line with wavelength of 532 nm (through fiber optics), as well as the computer equipped with dedicated software for precise laser control and accurate recording of SERS spectra. The two inlets are connected to the injection syringe pump. The detection of microorganisms takes place inside a microfluidic channel with an approximate width and height of 500 µm, carved by using a numerically controlled mechanical milling machine. Microorganisms are SERS detected inside the microfluidic cell, thus avoiding contamination. After passing through the microfluidic channel and when the bacteria detection process is completed, the biowaste is disposed through the outlet in a special container.
3.2. Synthesis of the Silver Spot Used for SERS-Based Detection of Bacteria
For the preparation of the SERS active substrate (see
Figure 2), we used the recipe published by Herman et al. [
5], with some modifications: While the spot was obtained by the authors of the article inside a glass capillary tube by injecting a mixture of the two components, in our case, we used a two-channel injection syringe pump that allows the simultaneous injection, with the same flow rate, of silver nitrate (AgNO
3) and sodium (Na) citrate inside the microchannel of the microfluidic cell.
3.3. The Detection Process of Microorganisms by Using SERS in the Microfluidic Cell
The detection process of microorganisms by using SERS takes place inside a microfluidic cell and uses an Ag spot synthesized in situ as enhancement substrate, under 532 nm laser irradiation. In short, the detection process comprises several steps:
Setting the flow parameters in the double syringes injection pump: Flow rate of 0.25 mL/min; syringe diameter: 12.97 mm; volume discharged: 5 mL.
Setting the laser power to 5 mW for the portable i-Raman BW-TEK spectrometer connected to the microscope by an optical fiber.
Concomitantly starting the laser and the injection syringe pump: the microfluidic channel is filled with the two components; local irradiation and heating by using the 532 nm laser Ag reduces to Ag0 and an Ag spot with plasmonic properties is created.
After the completion of the Ag spot generation process, the SERS substrate is tested by recording the bacteria specific spectra.
Recording SERS spectra and optimizing detection by testing the acquisition time and laser power appropriate in order to maintain the viability of the samples.Using a live CCD camera, the irradiated bacteria area was visualized in real time, prior, and during the spectral measurements.
4. Discussion
We consider that due to the fact that the Ag spot is formed on a solid surface (inside the microfluidic channel) and composed of Ag nanoclusters, the injected bacteria are adsorbed on the SERS substrate, due to the higher affinity of the cell wall components for the Ag nanoclusters than the existing citrate ions.
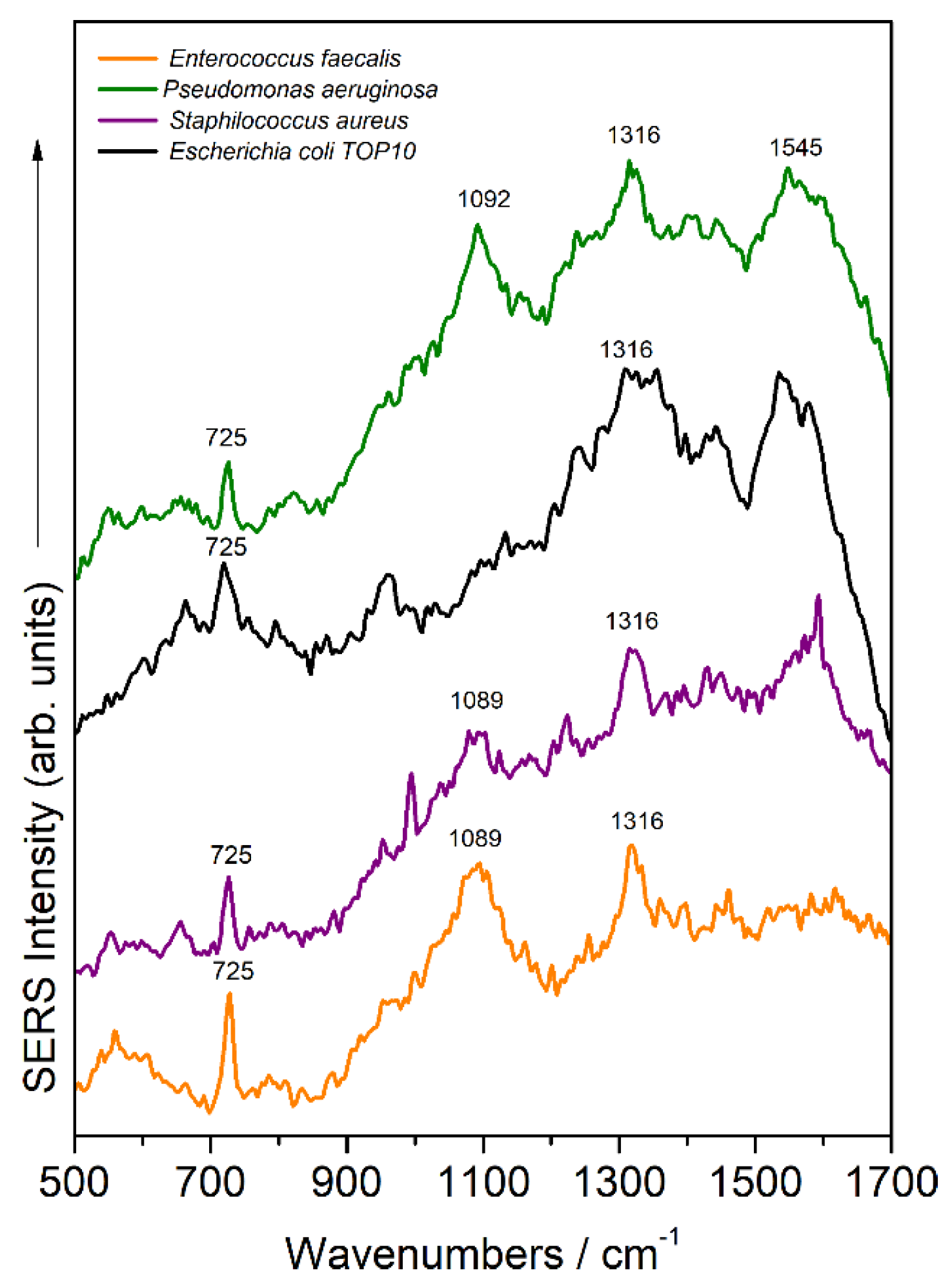
Figure 3 shows the SERS spectra of bacteria detected inside the microfluidic channel for four bacterial species. Both Gram-positive and Gram-negative species’ spectral fingerprints were successfully recorded. At least three of the SERS marker bands were detected for each species (725 cm
−1, 1089 cm
−1, and 1316 cm
−1, respectively), with slight Raman shifting from one species to another. These bands are assigned to living bacteria, since antibiotic stress or lysis is known to significantly affect [
6] the strong 725 cm
−1 and 1316 cm
−1 bands attributed to adenine and guanine, respectively, specific to SERS bacterial fingerprint [
7].
It is known that sample preparation for the SERS detection of bacteria is quite inconsistent referring to colloids as SERS active substrates. The NPs can be either coated on the outside of the bacterial cell wall or directed to the interior of the bacterial cells. Whereas the first preparation results in spectral information mainly derived from cell wall components and the second one contains additional cytoplasm information. By employing the herein described in situ preparation of silver SERS-active substrate, the spectral fingerprint obtained is from the cell wall of bacteria.
Conventional synthesis of colloidal suspension by using some reducing agents may result in the presence of extra ions originating from the reaction process [
8]. The instability of the sols has been one of our concerns and the presence of certain ions in the sols might be one reason for the unstable behavior of the sols. By using this preparation method, the presence of any byproducts and the instability of the nanoparticles are highly reduced. Additionally, spectral fluctuations deriving from the drying process are avoided and the spectral contributions recorded possess high specificity, arising for modes of flavin derivates prone to the SERS effect [
9].
Therefore, we believe that this is a promising technique for further relevant applications, wherever reliable bacteria validation is required.
5. Patents
A national patent registered at OSIM with nr. A00976/28.11.2018 resulting from the work reported in this manuscript.
Author Contributions
Conceptualization, N.E.D. and D.M.; methodology, A.C.; software and validation, A.M.R.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Romanian Ministry of Education (RME), CCCDI-UEFISCDI: grant numbers PN-III-P2-2.1-PED-2016-0983 (microfluidic device fabrication) and PN-III-P1-1.1-TE-2019-0910 (microfluidic device is still been used at the moment for SERS detection of bacteria).
Acknowledgments
The authors gratefully appreciate the help of PhD Tiberiu Szöke-Nagy from National Institute for Research and Development of Isotopic and Molecular Technologies, Molecular and Biomolecular Physics Department for providing the bacterial biomass in standardized samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ng, J.M.; Gitlin, I.; Stroock, A.D.; Whitesides, G.M. Components for integrated poly(dimethylsiloxane) microfluidic systems. Electrophoresis 2002, 23, 3461–3473. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Huang, C.-H.; Hsieh, W.-H.; Liu, L.-H.; Lin, Y.-C.; Chu, C.-C.; Wang, S.-T.; Kuo, I.-T.; Chau, L.-K.; Yang, C.-Y. On-line SERS Detection of Single Bacterium Using Novel SERS Nanoprobes and A Microfluidic Dielectrophoresis Device. Small 2014, 10, 4700–4710. [Google Scholar] [CrossRef]
- Chrimes, A.F.; Khoshmanesh, K.; Tang, S.-Y.; Wood, B.R.; Stoddart, P.R.; Collins, S.S.; Mitchell, A.; Kalantar-Zadeh, K. In situ SERS probing of nano-silver coated individual yeast cells. Biosens. Bioelectron. 2013, 49, 536–541. [Google Scholar] [CrossRef]
- Mungroo, N.A.; Oliveira, G.; Neethirajan, S. SERS based point-of-care detection of food-borne pathogens. Microchim. Acta 2015, 183, 697–707. [Google Scholar] [CrossRef]
- Herman, K.; Mircescu, N.E.; Szabo, L.; Leopold, L.F.; Chiş, V.; Leopold, N. In situ Silver Spot Preparation and on-Plate Surface-Enhanced Raman Scattering Detection in Thin Layer Chromatography Separation. J. Appl. Spectrosc. 2013, 80, 311–314. [Google Scholar] [CrossRef]
- Lemma, T.; Saliniemi, A.; Hynninen, V.; Hytönen, V.P.; Toppari, J.J. SERS detection of cell surface and intracellular components of microorganisms using nano-aggregated Ag substrate. Vib. Spectrosc. 2016, 83, 36–45. [Google Scholar] [CrossRef]
- Kang, J.W.; So, P.T.C.; Dasari, R.R.; Lim, D.-K. High Resolution Live Cell Raman Imaging Using Subcellular Organelle-Targeting SERS-Sensitive Gold Nanoparticles with Highly Narrow Intra-Nanogap. Nano Lett. 2015, 15, 1766–1772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, F.; Yin, H.; Hong, M. Nonuniform Distribution of Capping Ligands Promoting Aggregation of Silver Nanoparticles for Use as a Substrate for SERS. Adv. Nanoparticles 2013, 2, 104–111. [Google Scholar] [CrossRef][Green Version]
- Efrima, S.; Zeiri, L. Understanding SERS of bacteria. J. Raman Spectrosc. 2009, 40, 277–288. [Google Scholar] [CrossRef]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).