Identification of Prevotella, Anaerotruncus and Eubacterium Genera by Machine Learning Analysis of Metagenomic Profiles for Stratification of Patients Affected by Type I Diabetes †
Abstract
:1. Introduction
2. Materials and Methods
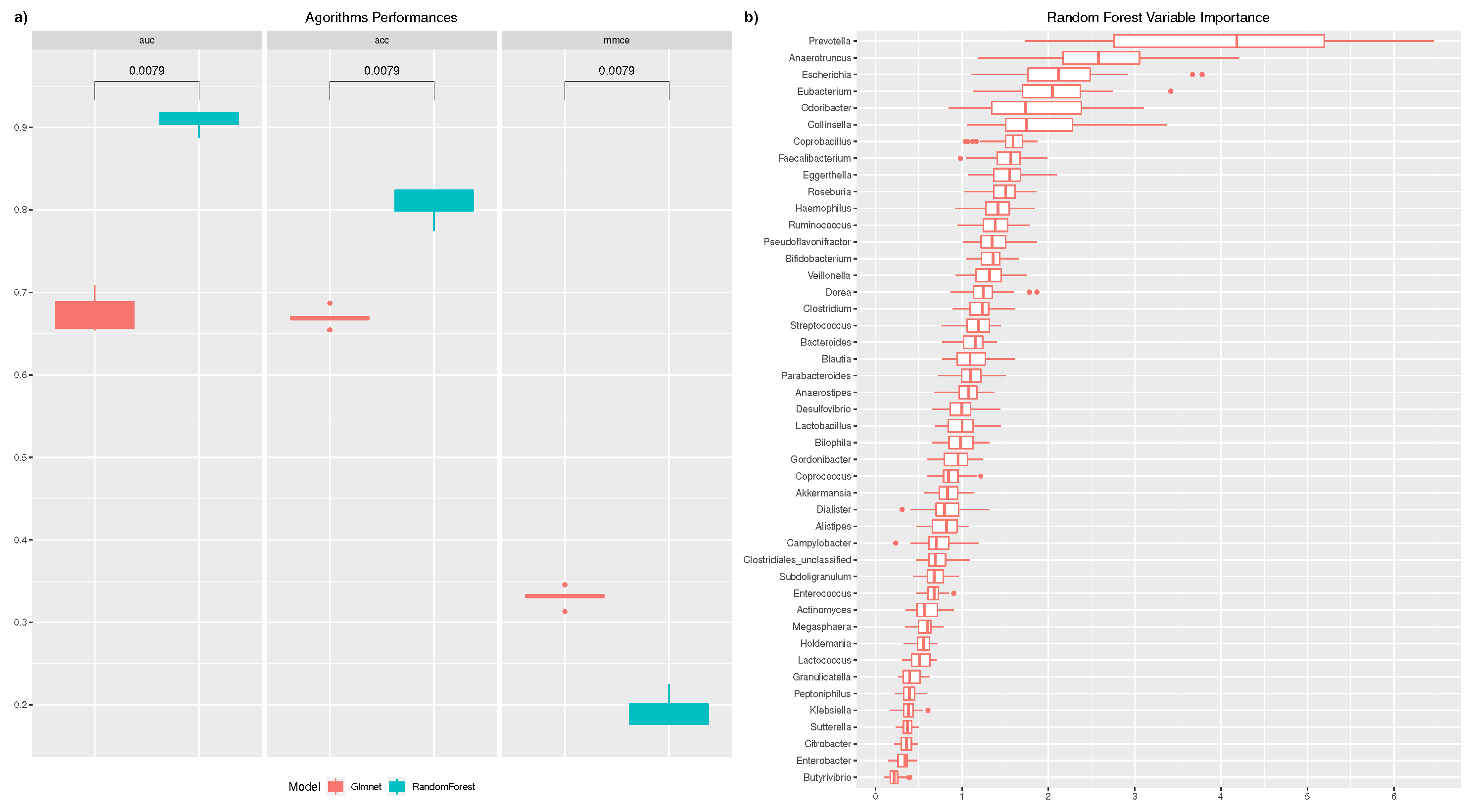
3. Results
4. Discussion
Author Contributions
Funding
References
- Tai, N.; Wong, F.S.; Wen, L. The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Rev. Endocr. Metab. Disord. 2015, 16, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyötyläinen, T.; Hämäläinen, A.M.; Peet, A.; Tillmann, V.; Pöhö, P.; Mattila, I.; et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell Host Microbe 2016, 20, 121. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Siljander, H.; Honkanen, J.; Knip, M. Microbiome and type 1 diabetes. EBioMedicine 2019, 46, 512–521. [Google Scholar] [CrossRef]
- Hasan, S.; Aho, V.; Pereira, P.; Paulin, L.; Koivusalo, S.B.; Auvinen, P.; Eriksson, J.G. Gut microbiome in gestational diabetes: a cross-sectional study of mothers and offspring 5 years postpartum. Acta Obstet. Gynecol. Scand. 2018, 97, 38–46. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Edreira, D.; Liñares-Blanco, J.; Fernandez-Lozano, C. Identification of Prevotella, Anaerotruncus and Eubacterium Genera by Machine Learning Analysis of Metagenomic Profiles for Stratification of Patients Affected by Type I Diabetes. Proceedings 2020, 54, 50. https://doi.org/10.3390/proceedings2020054050
Fernández-Edreira D, Liñares-Blanco J, Fernandez-Lozano C. Identification of Prevotella, Anaerotruncus and Eubacterium Genera by Machine Learning Analysis of Metagenomic Profiles for Stratification of Patients Affected by Type I Diabetes. Proceedings. 2020; 54(1):50. https://doi.org/10.3390/proceedings2020054050
Chicago/Turabian StyleFernández-Edreira, Diego, Jose Liñares-Blanco, and Carlos Fernandez-Lozano. 2020. "Identification of Prevotella, Anaerotruncus and Eubacterium Genera by Machine Learning Analysis of Metagenomic Profiles for Stratification of Patients Affected by Type I Diabetes" Proceedings 54, no. 1: 50. https://doi.org/10.3390/proceedings2020054050
APA StyleFernández-Edreira, D., Liñares-Blanco, J., & Fernandez-Lozano, C. (2020). Identification of Prevotella, Anaerotruncus and Eubacterium Genera by Machine Learning Analysis of Metagenomic Profiles for Stratification of Patients Affected by Type I Diabetes. Proceedings, 54(1), 50. https://doi.org/10.3390/proceedings2020054050





