Paired and Unpaired Deep Generative Models on Multimodal Retinal Image Reconstruction †
Abstract
:1. Introduction
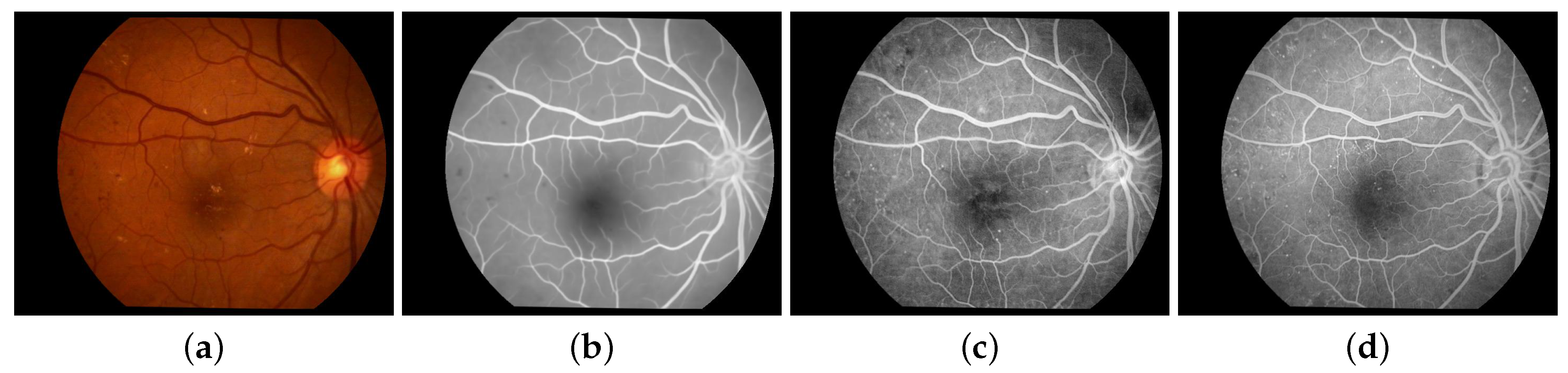
2. Methodology
3. Results and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hervella, A.S.; Rouco, J.; Novo, J.; Ortega, M. Retinal Image Understanding Emerges from Self-Supervised Multimodal Reconstruction. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention (MICCAI), Granada, Spain, 16–20 September 2018. [Google Scholar]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Hervella, A.S.; Rouco, J.; Novo, J.; Ortega, M. Multimodal Registration of Retinal Images Using Domain-Specific Landmarks and Vessel Enhancement. In Proceedings of the International Conference on Knowledge-Based and Intelligent Information and Engineering Systems (KES), Belgrade, Serbia, 3–5 September 2018. [Google Scholar]
- Hervella, A.S.; Rouco, J.; Novo, J.; Ortega, M. Deep Multimodal Reconstruction of Retinal Images Using Paired or Unpaired Data. In Proceedings of the International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hervella, Á.; Rouco, J.; Novo, J.; Ortega, M. Paired and Unpaired Deep Generative Models on Multimodal Retinal Image Reconstruction. Proceedings 2019, 21, 45. https://doi.org/10.3390/proceedings2019021045
Hervella Á, Rouco J, Novo J, Ortega M. Paired and Unpaired Deep Generative Models on Multimodal Retinal Image Reconstruction. Proceedings. 2019; 21(1):45. https://doi.org/10.3390/proceedings2019021045
Chicago/Turabian StyleHervella, Álvaro, José Rouco, Jorge Novo, and Marcos Ortega. 2019. "Paired and Unpaired Deep Generative Models on Multimodal Retinal Image Reconstruction" Proceedings 21, no. 1: 45. https://doi.org/10.3390/proceedings2019021045
APA StyleHervella, Á., Rouco, J., Novo, J., & Ortega, M. (2019). Paired and Unpaired Deep Generative Models on Multimodal Retinal Image Reconstruction. Proceedings, 21(1), 45. https://doi.org/10.3390/proceedings2019021045






