Abstract
Peptide cross-linked poly(ethylene glycol) hydrogel is a promising biomaterial that has been used widely for drug delivery and tissue engineering. However, the use of this material as a biosensor material for the detection of collagenase has not been explored. Collagenase play a key role in rheumatoid arthritis and osteoarthritis. Detection of this class of enzyme using the degradable hydrogel film format is promising as a point-of-care device for disease monitoring. In this study, a biosensor was developed based on the degradation of a peptide cross-linked poly(ethylene glycol) hydrogel film for the detection of collagenase. The hydrogel was deposited on gold-coated quartz crystals and their degradation in the presence of collagenase was monitored using a Quartz Crystal Microbalance (QCM). The biosensor was shown to respond to concentrations between 2 nM to 2000 nM with a lower detection limit of 2 nM.
1. Introduction
Biosensors based on the degradation of hydrogel films have previously been introduced for the detection of various proteases. The degradation of peptide cross-linked dextran hydrogel films was successfully monitored using QCM and impedance measurements at interdigitated electrodes in the presences of periodontal disease biomarkers: human neutrophil elastase (HNE), cathepsin G and matrix metalloproteinase-8 (MMP-8) [1,2]. In both studies, different peptide cross-linkers were used and the results show high specificity of the biosensor to the enzyme of interest. Collagenase levels were previously reported to be higher in synovial fluid of rheumatoid arthritis (RA) and osteoarthritis (OS) patients, and correlated with inflammation in acute RA and chronic pathology of OS [3].
Poly(ethylene glycol) (PEG) is a hydrophilic polymer and has been reported to have anti-fouling properties [4]. The use of dendritic architecture of PEG as biosensor material is promising as it has more active sites for cross-linking and low non-specific binding. The cross-linking of 4-arm PEG norbornene with specific peptide sequences is likely to offer good sensor responses and high specificity based on the peptide cross-linker used. In this study, the peptide cross-linker, GCRDVPMS↓MRGGDRCG was used to cross-link 4-arm PEG norbornene (MW 20,000) to detect collagenase. A solution containing the peptide and polymer in PBS pH 6 was drop coated onto a hydrophobic glass slide. The QCM crystal was placed on top of the solution. The hydrogel formed after UV exposure for 300 s. The degradation of peptide cross-linked PEG was detected in the presence of collagenase using QCM measurements (Scheme 1). The sensor was shown to be sensitive in a concentration range of 2 nM to 2000 nM.
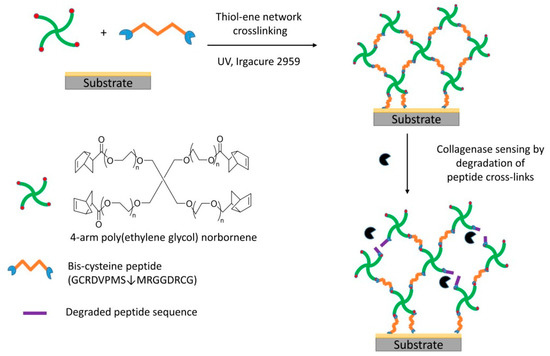
Scheme 1.
Sensor fabrication and collagenase sensing on gold coated QCM crystals.
2. Materials and Methods
2.1. Materials
Custom peptide GCRDVPMS↓MRGGDRCG (↓ shows cleavage site, MW 1696.97 Da) was purchased from Proteogenix, Schiltigheim, France. Collagenase from Clostridium histolyticum for general used Type I (approximate MW 125,000 Da), 4-arm poly(ethylene glycol) norbornene (MW 20,000), tricine (N-[tris(hydroxymethyl)methyl]glycine, 99%), sodium chloride (99.5%), calcium chloride dihydrate (99%), phosphate buffer saline tablets, anhydrous toluene (99.8%) and trichloro(octadecyl)silane (90%) were purchased from Sigma. Concentrated sulfuric acid (95–98%), zinc chloride and hydrogen peroxide (30% H2O2 (w/w)) were purchased from Fluka. Sodium hydroxide (1 M) was purchased from Fisher Chemicals, UK. Microscopic glass slides (Menzel-glaser, 75 mm × 25 mm) from Thermo Scientific were used in sensor fabrication. Polished, gold coated QCM crystals (AT-cut, 10 MHz) were purchased from International Crystal Manufacturing Company, Inc. (Oklahoma City, OK, USA). All solutions were prepared with Milli-Q water (resistivity 18.2 MΩ.cm).
2.2. Sensor Fabrication
Prior to the sensor fabrication, 200 µL of hydrogel solution in PBS pH 6 for 100% cross-linking was prepared by mixing 104.2 µL of PEG NB (192.4 mg/mL) and 17.8 µL of peptide cross-linker (GCRDVPMS↓MRGGDRCG) (190.9 mg/mL). 5 mol % of the photo-initiator Irgacure 2959 in methanol was added last and the mixture was kept away from sunlight and used freshly. To silanize the glass slides, they were immersed in 0.2% tricholoro(octadecyl)silane in anhydrous toluene for 90 min and kept dry after rinsing.
For the sensor fabrication, the hydrogel solution was sandwiched between a silanized glass slide and a QCM crystal and exposed to UV light (17 mW/cm2, 350–500 nm) for 300 s using a UV lamp Omnicure series 1500 [5]. The coated QCM crystal was then removed from the silanized glass slide and kept in PBS buffer before use. Scheme 1 shows the schematic diagram of sensor fabrication and collagenase sensing.
2.3. Monitoring Degradation of Hydrogel Using QCM Measurements
Degradation of the hydrogel by collagenase was monitored using QCM measurement. The QCM data was analysed using the Butterworth Van Dyke (BVD) equivalent circuit with ΔR representing the viscoelasticity and ωΔL representing the mass of the hydrogel on the gold coated QCM crystal [6]. Prior to the degradation experiment, the hydrogel was first equilibrated in PBS buffer pH 7 before exposure to collagenase. Varying concentrations of collagenase (0.2–2000 nM) were used to investigate the effect of enzyme concentration on the degradation of the hydrogel with degradation recorded for 10 min after exposure to collagenase.
3. Results and Discussion
Degradation of the peptide cross-linked PEG hydrogel was investigated using QCM in PBS buffer and was exposed to 20 nM collagenase over 10 min as shown in Figure 1. The sensor response was stable in PBS buffer before exposure to the collagenase. A significant decrease of QCM response, ωΔL and ΔR were observed after exposure to collagenase, which corresponds to the cleavage of the peptide cross-linker in the hydrogel films. The data shows that ΔR was more significant than ωΔL in the QCM response, thus ΔR will be used in the further discussion.
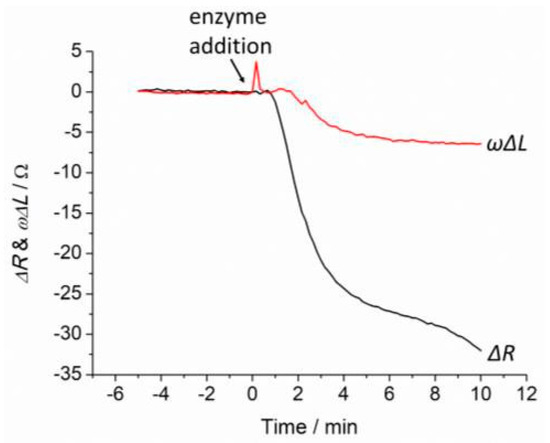
Figure 1.
QCM response, ΔR and ωΔL of 100% GCRDVPMS↓MRGGDRCG cross-linked PEG after addition of 20 nM over 10 min.
Figure 2 shows the response of 100% GCRDVPMS↓MRGGDRCG cross-linked PEG hydrogel to different concentrations of collagenase in a range of 0.2 nM to 2000 nM. From Figure 2a, the sensor successfully detected collagenase and was shown to be concentration dependent with a lower detection limit of 2 nM. An increase of the sensor response, ΔR, was observed as the concentration of collagenase increased (Figure 2a), which corresponds to a faster degradation of the peptide cross- links with increasing collagenase concentration. At high concentrations of collagenase, the hydrogel films were observed to be smoother after 10 min exposure to the enzyme compared to a rough surface before the degradation. A collagenase concentration of 0.2 nM showed no response which indicated that no significant degradation of the hydrogel occurred.
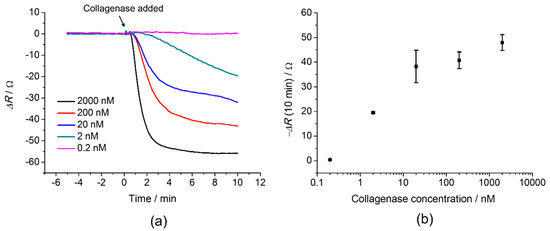
Figure 2.
(a) QCM response, ΔR of 100% GCRDVPMS↓MRGGDRCG cross-linked PEG after addition of 0.2 nM, 2 nM, 20 nM, 200 nM and 2000 nM at t = 0. (b) Concentration dependence of the QCM response, average of ΔR, measured for 10 min after the addition of collagenase.
4. Conclusions
Here, a collagenase biosensor was successfully developed. The sensor shows a QCM response dependent on collagenase concentration, which was tested within a range of 0.2 nM to 2000 nM. The lower detection limit of this sensor was 2 nM of collagenase. Further investigation of the effect of the degree of cross-linking on the degradation by collagenase is currently under way.
Author Contributions
N.A. performed the experiments and wrote the paper, B.C. performed experiments and co-wrote the paper, M.J.G., D.-W.Z., M.W., C.R.B., J.E.G. and S.K. contributed to the experimental design and co-wrote the paper.
Acknowledgments
The authors gratefully acknowledge the Ministry of Higher Education, Malaysia, for providing a PhD studentship to N.A. The authors are grateful to the Biotechnology and Biological Sciences Research Council (BBSRC) of UK for providing research funding (BB/ P026788/1) and to the EU for providing a Marie Skłodowska-Curie Individual Fellowship (H2020-MSCA-IF-2014-660489).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Biela, A.; Watkinson, M.; Meier, U.C.; Baker, D.; Giovannoni, G.; Becer, C.R.; Krause, S. Disposable MMP-9 sensor based on the degradation of peptide cross-linked hydrogel films using electrochemical impedance spectroscopy. Biosens. Bioelectron. 2015, 68, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Cook, J.P.; Watkinson, M.; Yang, S.; Douglas, I.; Rawlinson, A.; Krause, S. Generic protease detection technology for monitoring periodontal disease. Faraday Discuss. 2011, 149, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Clark, I.M.; Powell, L.K.; Ramsey, S.; Hazleman, B.L.; Cawston, T.E. The measurement of collagenase, tissue inhibitor of metalloproteinases (timp), and collagenase—Timp complex in synovial fluids from patients with osteoarthritis and rheumatoid arthritis. Arthritis Rheum. 1993, 36, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, I.; Pangule, R.C.; Kane, R.S. Antifouling coatings: Recent developments in the design of surfaces that prevent fouling by proteins, bacteria, and marine organisms. Adv. Mater. 2011, 23, 690–718. [Google Scholar] [CrossRef] [PubMed]
- Colak, B.; Da Silva, J.C.S.; Soares, T.A.; Gautrot, J.E. Impact of the Molecular Environment on Thiol- Ene Coupling for Biofunctionalization and Conjugation. Bioconjug. Chem. 2016, 27, 2111–2123. [Google Scholar] [CrossRef] [PubMed]
- Sabot, A.; Krause, S. Simultaneous quartz crystal microbalance impedance and electrochemical impedance measurements. Investigation into the degradation of thin polymer films. Anal. Chem. 2002, 74, 3304–3311. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
