1. Introduction
The on-site, readily detection of hazardous bacteria varieties by means of on-chip microfluidic systems has become a relevant trend in the development of Lab-on-Chip and Micro Analysis Devices [
1], given their reduced analysis time and cost in comparison to traditional off-chip analytic assay methods [
2]. Typically, the manufacturing of micro analysis devices on silicon and glass substrates have favored their miniaturization and integration with sensors and actuators. Nevertheless, the reduction of production costs as well as the compliance to biosafety regulations to use them in clinical and environmental assays, have motivated the development of microfluidic biosensors of disposable materials [
3], which in turn impose important challenges such as the integration of the diverse capabilities necessary to promote the biorecognition reactions on a single microfluidic device. In order to address such challenges, micro analysis devices have been widely developed on thermoplastic substrates. Among them, Cyclo-Olefin Polymer (COP) and Cyclo-Olefin Copolymer (COC) materials have earned great acceptance due to their low water absorption, low auto fluorescence and high chemical resistance [
4]. Injection molding and hot embossing make possible to structure features the range of microns over such thermoplastic substrates.
The present work describes the design and prototyping of a disposable microfluidic biosensor, which is fully made of COC and performs bacteria heat lysis for RNA retrieval, sample cooling and analysis reaction on the same substrate. The device is tested as microfluidic biosensor of E. coli based on RNA hybridization and fluorescence detection. The observed performance proves the potential application of a disposable microfluidic biosensor at real-time, lab-on-chip based monitoring and control of bacteria pollution.
2. Materials and Methods
2.1. Biosensor Design and Preparation
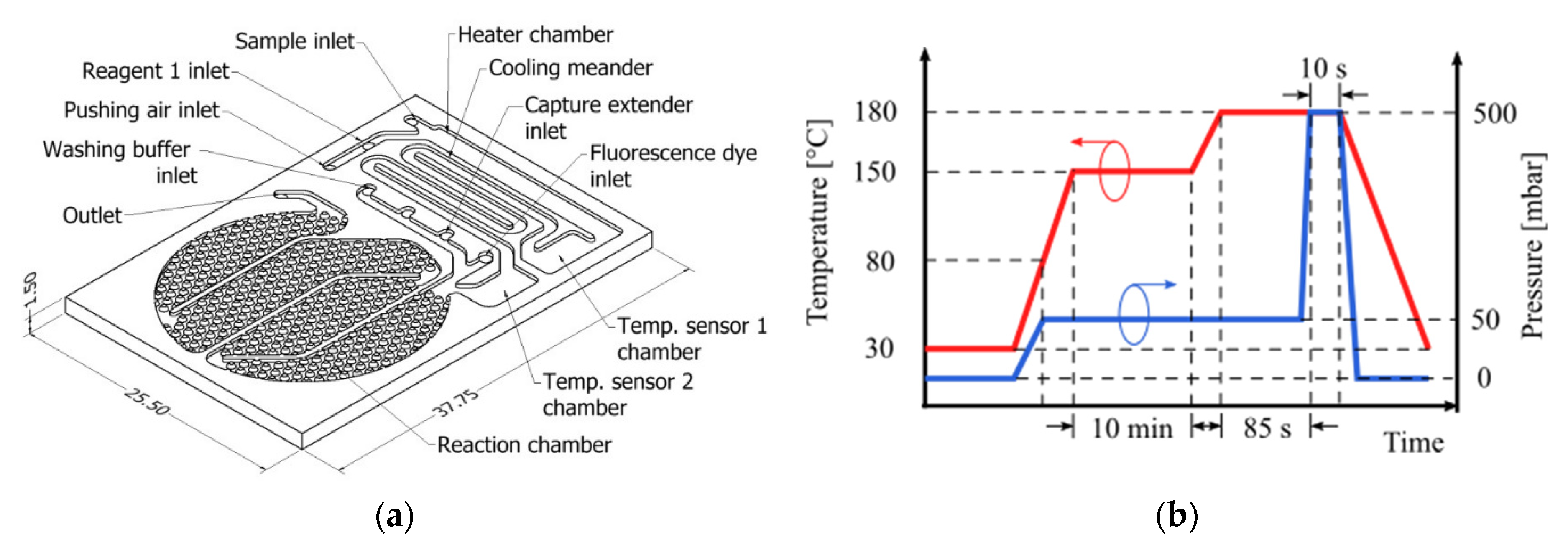
The microfludic biosensor was designed to perform sample heating, cooling and detection reaction (total fluid volume 134 µL). In addition, it should allow on-process sample temperature measurements and injection of the reagents participating in the detection reaction. The design concept is illustrated in
Figure 1a. The surface on the reaction chamber (75 µL) has been patterned with obstacles in order to increase the surface-to-volume ratio and promote adsorption on the surface activated with reactive molecules [
5]. The round shape of the reaction chamber was designed to fit under a 1″ diameter optical fitting. Partition walls in the reaction chamber aims to maximize the fluid sample transit time over the activated surface and by this way improve the occurrence of binding events and thus the biosensor effectivity. The channels, as well as the chambers, deepen to 300 µm. Before manufacturing rheological characterization of COC was conducted in order to estimate the material viscosity vs. temperature profile. From these results, the hot embossing process parameters were determined and applies as indicated in
Figure 1b. COC plates, from TOPAS grade 5013 (transition temperature T
g of 142 °C), were processed by hot embossing to structure the substrate.
Before closing the microfluidic structures, the reaction chamber surface was activated with capture molecules, using a customized technique based on the method described in [
6]. Silver heater and carbon temperature sensitive resistors were screen-printed on a 100 µm thickness, T
g = 142 °C, COC TOPAS 5013 grade foil, in the way the printed elements match the heater chamber and temperature sensor layouts. The printed side of the foil was hot pressed against a COC TOPAS 8007 foil T
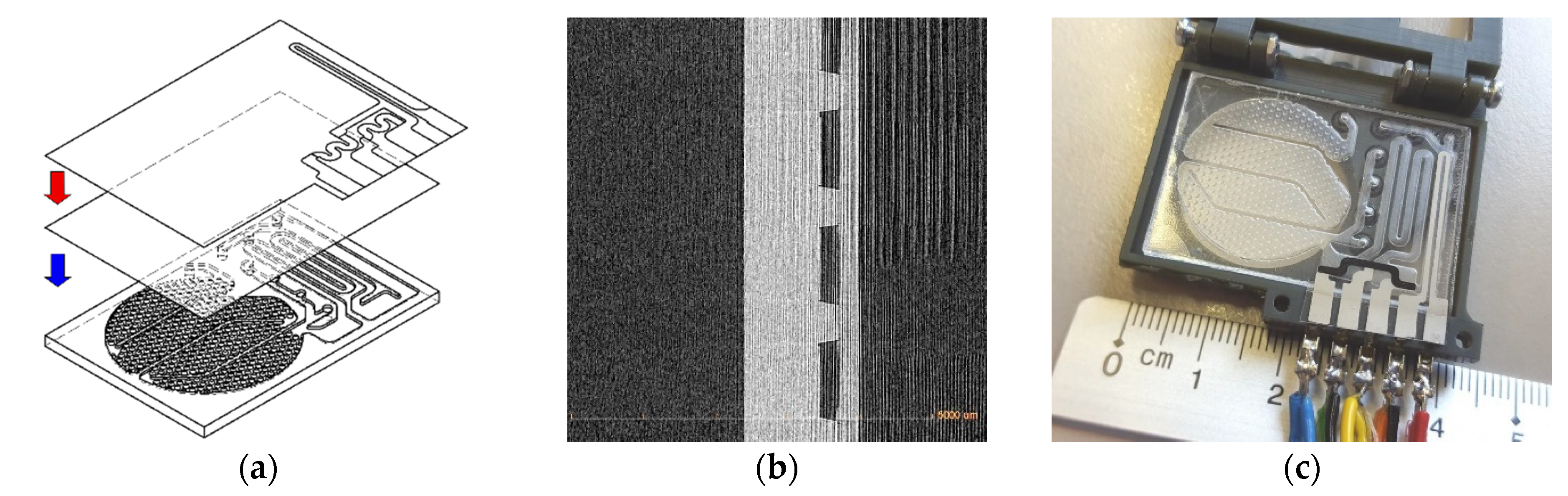
g = 78 °C, applying 110 °C, 500 mbar pressure and vacuum. Later the compound layer is aligned over the substrate layout and hot pressed on it (
Figure 2a), by applying 100 °C from the bottom plate and 500 mbar pressure. An X-ray cross section image of the resulting closed microchannels is shown in
Figure 2b. A prepared biosensor is shown in
Figure 2c, including a screen printed heater and temperature sensitive resistances.
2.2. Experimental Setup
A 1 mL volume sample is injected into the biosensor, flowing through the activated surface at a rate of 14 mL min−1 (off-chip processed RNA, diluted bacteria and negative control). An external heat source keeps the COC substrate material at 50 °C to temperate reaction chamber to a favorable temperature for RNA hybridization. A customized buffer was prepared to be injected after every hybridization step and wash off unbound RNA, and to be beforehand mixed with the sample in case of the 95 °C on-chip heat lysis, in order to improve the quality of retrieved RNA. Bound molecules were tagged with DY-480XL fluorescence dye and detected using a photon counter.
3. Results
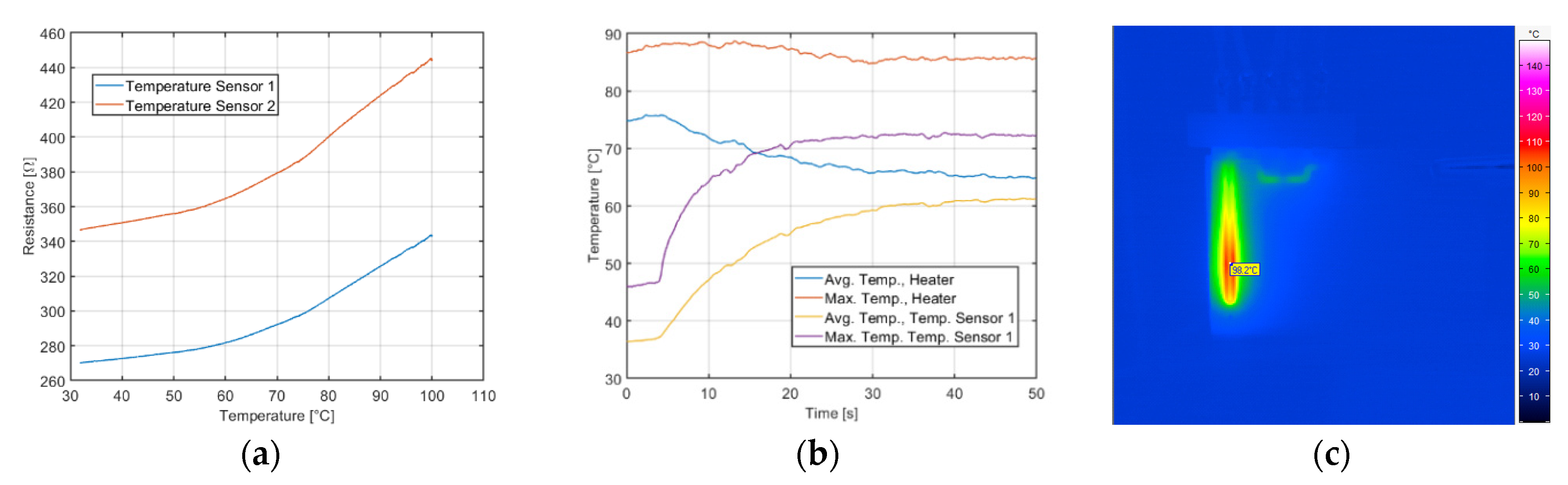
The functionality of printed heater and temperature sensors were tested before closing the microfluidic devices. The printed carbon resistor on COC foils exhibited increasing resistance at increasing temperature, resulting in positive temperature coefficients (TCR). Resistance vs. temperature profiles showed a close approximation to exponential functions in most of the cases as shown as example in
Figure 3a. Average resistances and average temperature coefficients for temperature sensors 1 and 2 are 363.5 Ω and 365.4 Ω respectively. Silver heater printed structured yielded an average resistance of 7.9 Ω. After closing the microfluidic device, heater was tested with water sample and temperature recorded by mean of thermal infrared (IR) imaging, whose temperature time-profile is shown in
Figure 3b.
Figure 3c shows the temperature spatial distribution over the device with active heater and no flow.
On-chip heat-lysis function was tested by flushing sample water with
E. coli concentration applying heater temperature around 95 °C and lately analyzing on PCR the quality of the released RNA. The RNA hybridization on the activated detection surface and fluorescence detection was conducted following a proprietary procedure created for this development, whose fundamentals are based on the method proposed in [
6]. On-chip fluorescence quantitative results showed an off-chip processed RNA detection signal intensity 30.4% higher than background signal, and an on-chip retrieved RNA signal intensity 4.3% higher than background signal.
4. Discussion and Conclusions
The COC rheological characterization results proved to be critical for the determination of the hot embossing parameters. Moreover the characterization results offer the potentiality to estimate parameters for different geometries and feature sizes, mostly imposed by the mold manufacturing possibilities. On the other hand, the employed foil sealing technique consisting on hot pressing applying temperature from bottom substrate side proved to obtain better resulting channel integrity in comparison to apply the molding heat from the foil side, arguably own to a selective bonding heat distribution only on the contact area between COC foil and COC substrate. Material qualities such as low auto-fluorescence and biocompatibility could be of benefit to this fluorescence detection based biosensor. Nevertheless, the application of screen printed sensors still rendered limitations regarding accuracy and reproducibility, so the refinement of their implementation technique on COC foils is advised.
On-chip RNA hybridization proved to work by applying off-chip processed RNA and detecting the binding events over the reaction surface by fluorescence signal, suggesting that the applied functionalization technique of the COC surface can consistently perform for hybridization based biosensors. Nonetheless, the dim detected fluorescence intensity after the on-chip heat-lysed bacteria RNA retrieval suggests that, although a positive detection reading was observed, more results are needed to estimate the dynamic range and sensitivity of the biosensor. The effect of variables such as the sample injection flow rate and the observed formation of foam in the hybridization buffer probably impacts negatively the hybridization reaction efficiency. The study and control of this variables were out of the scope of this work, and its conduction on the designed device is a motivation for future works.
As proof of concept, this work showed that RNA based bacteria detection can be implemented on a single substrate, disposable material, microfluidic chip, suggesting its potential for further customization and optimization for detection of another pathogenic bacteria species.