Sensor Array Based on Molecularly Imprinted Polymers for Simultaneous Detection of Lipoproteins †
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Whitcombe, M.J.; Chianella, I.; Larcombe, L.; Piletsky, S.A.; Noble, J.; Porter, R.; Horgan, A. The rational development of molecularly imprinted polymer-based sensors for protein detection. Chem. Soc. Rev. 2011, 40, 1547–1571. [Google Scholar] [CrossRef] [PubMed]
- Cieplak, M.; Szwabinska, K.; Sosnowska, M.; Chandra, B.K.C.; Borowicz, P.; Noworyta, K.; D’Souza, F.; Kutner, W. Selective electrochemical sensing of human serum albumin by semi-covalent molecular imprinting. Biosens. Bioelectron. 2015, 74, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Ren, L.; Zhao, H.; Xu, C.; Zhang, L.; Yu, Y.; Wang, H.; Lan, Y.; Roberts, M.F.; Chuang, J.H.; et al. A molecular-imprint nanosensor for ultrasensitive detection of proteins. Nat. Nanotechnol. 2010, 5, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Schirhagl, R.; Podlipna, D.; Lieberzeit, P.A.; Dickert, F.L. Comparing biomimetic and biological receptors for insulin sensing. Chem. Commun. 2010, 46, 3128–3130. [Google Scholar] [CrossRef] [PubMed]
- Barter, P.J.; Rye, K.A. High density lipoproteins and coronary heart disease. Atherosclerosis 1996, 121, 1–12. [Google Scholar] [CrossRef]
- Nauck, M.; Warnick, G.R.; Rifai, N. Methods for measurement of ldl-cholesterol: A critical assessment of direct measurement by homogeneous assays versus calculation. Clin. Chem. 2002, 48, 236. [Google Scholar] [CrossRef] [PubMed]
- Chunta, S.; Suedee, R.; Lieberzeit, P.A. Low-density lipoprotein sensor based on molecularly imprinted polymer. Anal. Chem. 2016, 88, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Chunta, S.; Suedee, R.; Lieberzeit, P.A. High-density lipoprotein sensor based on molecularly imprinted polymer. Anal. Bioanal. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Dickert, F.; Hayden, O.; Lieberzeit, P.; Palfinger, C.; Pickert, D.; Wolff, U.; Scholl, G. Borderline applications of qcm-devices: Synthetic antibodies for analytes in both nm- and μm-dimensions. Sens. Actuators B 2003, 95, 20–24. [Google Scholar] [CrossRef]
- Lieberzeit, P.A.; Schirk, C.; Glanznig, G.; Gazda-Miarecka, S.; Bindeus, R.; Nannen, H.; Kauling, J.; Dickert, F.L. From nanopatterning to functionality—Surface and bulk imprinting for analytical purposes. Superlattices Microstruct. 2004, 36, 133–142. [Google Scholar] [CrossRef]
- Shen, B.W.; Scanu, A.M.; Kézdy, F.J. Structure of human serum lipoproteins inferred from compositional analysis. Proc. Natl. Acad. Sci. USA 1977, 74, 837–841. [Google Scholar] [CrossRef] [PubMed]
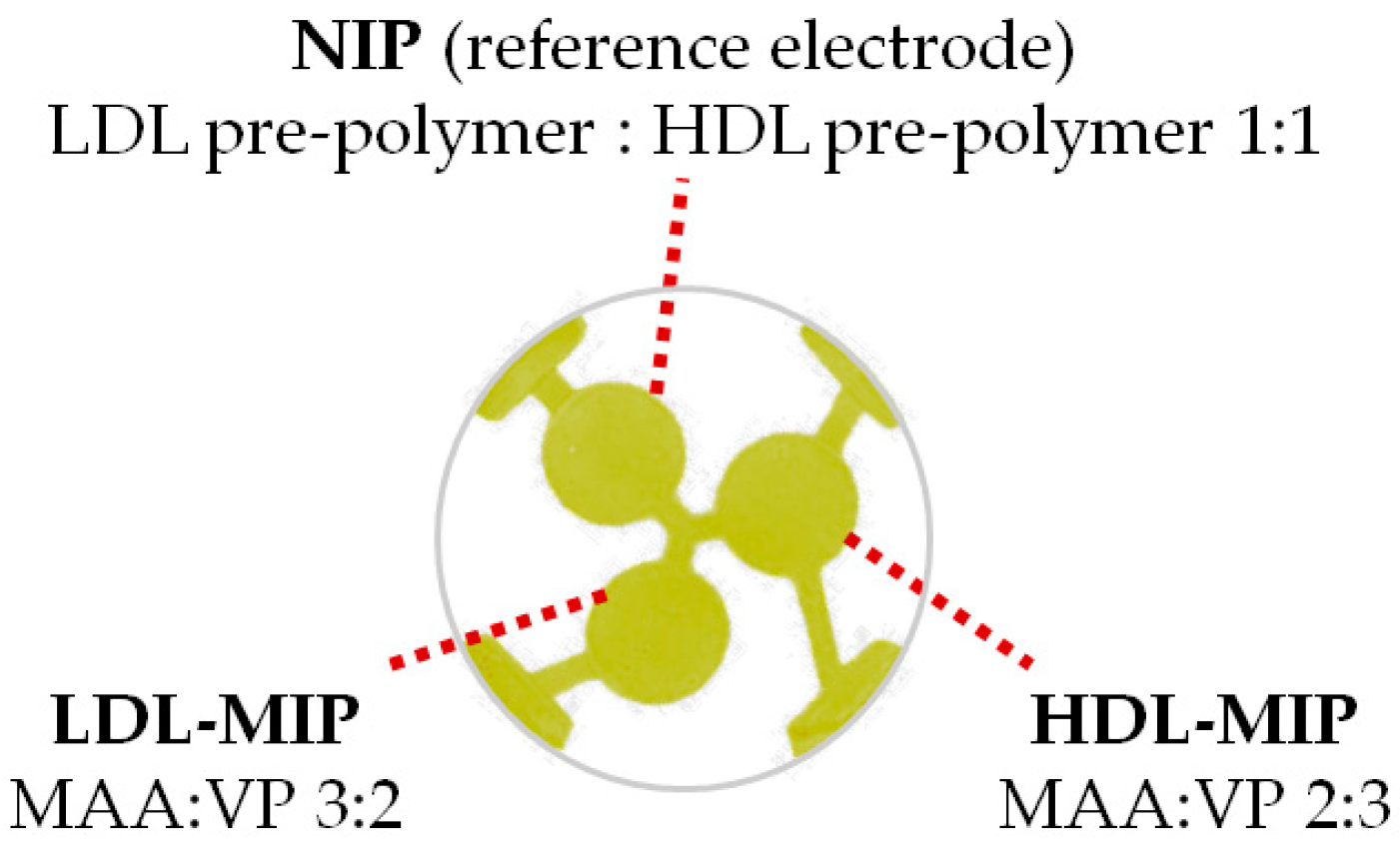
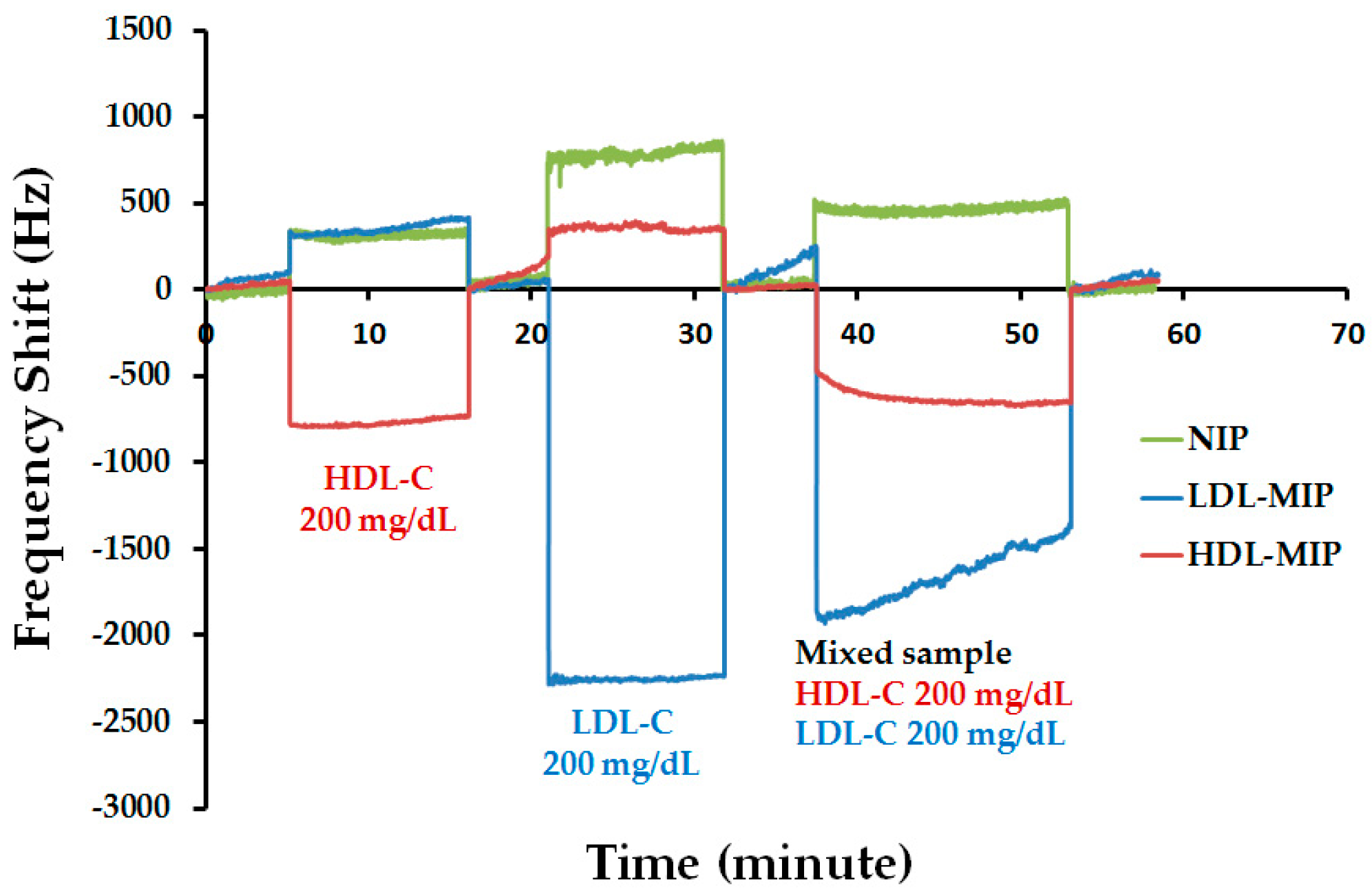
| Risk Level of CHD | LDL-C (mg/dL) | HDL-C (mg/dL) |
|---|---|---|
| Low risk | <129 | ≥60 |
| High risk | 130–159 | <40 |
| Very high risk | >159 | - |
| Mixed-Standard Concentration | Mean of Frequency Shift (Hz) (n = 3) | Standard Deviation | % CV |
|---|---|---|---|
| 200 mg/dL LDL-C | 1619 | 39 | 2 |
| 100 mg/dL HDL-C | 471 | 37 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chunta, S.; Singsanan, S.; Suedee, R.; Lieberzeit, P.A. Sensor Array Based on Molecularly Imprinted Polymers for Simultaneous Detection of Lipoproteins. Proceedings 2017, 1, 510. https://doi.org/10.3390/proceedings1040510
Chunta S, Singsanan S, Suedee R, Lieberzeit PA. Sensor Array Based on Molecularly Imprinted Polymers for Simultaneous Detection of Lipoproteins. Proceedings. 2017; 1(4):510. https://doi.org/10.3390/proceedings1040510
Chicago/Turabian StyleChunta, Suticha, Sanita Singsanan, Roongnapa Suedee, and Peter A. Lieberzeit. 2017. "Sensor Array Based on Molecularly Imprinted Polymers for Simultaneous Detection of Lipoproteins" Proceedings 1, no. 4: 510. https://doi.org/10.3390/proceedings1040510
APA StyleChunta, S., Singsanan, S., Suedee, R., & Lieberzeit, P. A. (2017). Sensor Array Based on Molecularly Imprinted Polymers for Simultaneous Detection of Lipoproteins. Proceedings, 1(4), 510. https://doi.org/10.3390/proceedings1040510





