Harnessing Legume Productivity in Tropical Farming Systems by Addressing Challenges Posed by Legume Diseases
Abstract
1. Introduction
2. What Is the Ecological and Economic Importance of Legumes?
3. What Are the Key Diseases Affecting Legumes in Tropical Farming Systems?
3.1. Bacterial Diseases
3.2. Fungal Diseases
3.3. Oomycete Diseases
3.4. Viral Diseases
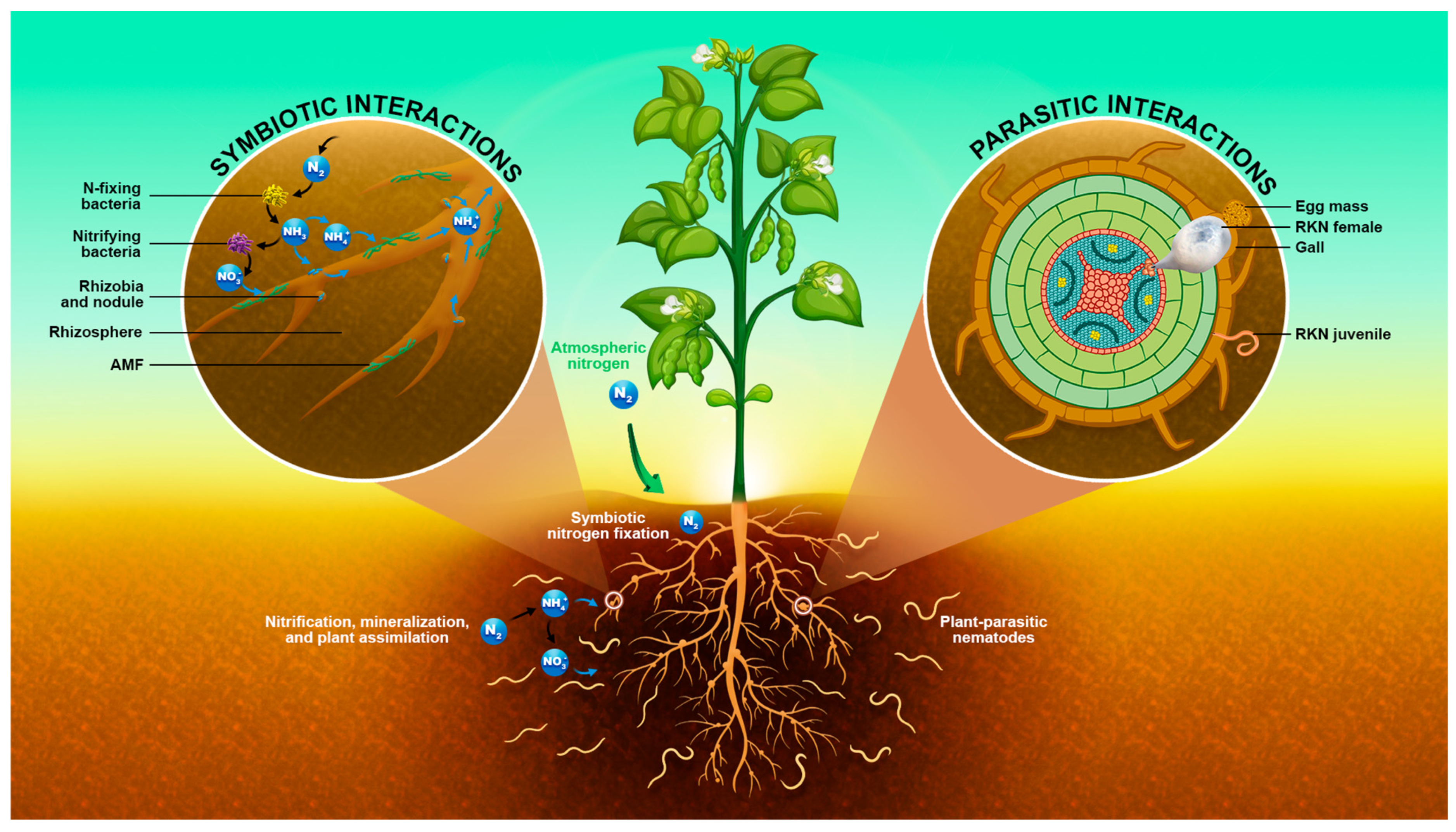
3.5. Nematode Diseases and Nematode-Rhizosphere Interaction
3.6. How Do Rhizobia, the Plant’s Physiology, and Soil Nutrition Influence Disease Development in Legumes?
4. What Are the Economic, Environmental, and Social Consequences of Legume Diseases?
4.1. Economic Consequences
4.2. Environmental Impact
4.3. Social Implications: Livelihoods of Smallholder Farmers, Food Security Concerns
5. Tackling the Challenges Associated with Legume Diseases: The Way Forward
5.1. Harnessing Utilization of Legume Genetic Resources for Disease Resistance
5.2. Innovative Breeding and Biotechnology Advancements in Legumes in the Tropics
5.3. PGR Contributions to Smallholder Resilience and Food Security
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Morales, F.J. Common Beans. In Natural Resistance Mechanisms of Plants to Viruses; Loebenstein, G., Carr, J.P., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 367–382. [Google Scholar]
- Rodríguez Madrera, R.; Campa Negrillo, A.; Ferreira Fernández, J.J. Modulation of the Nutritional and Functional Values of Common Bean by Farming System: Organic vs. Conventional. Front. Sustain. Food Syst. 2024, 7, 1282427. [Google Scholar] [CrossRef]
- Gondwe, T.M.; Alamu, E.O.; Mdziniso, P.; Maziya-Dixon, B. Cowpea (Vigna unguiculata (L.) Walp) for Food Security: An Evaluation of End-User Traits of Improved Varieties in Swaziland. Sci. Rep. 2019, 9, 15991. [Google Scholar] [CrossRef]
- Togola, A.; Datinon, B.; Laouali, A.; Traoré, F.; Agboton, C.; Ongom, P.O.; Ojo, J.A.; Pittendrigh, B.; Boukar, O.; Tamò, M. Recent Advances in Cowpea IPM in West Africa. Front. Agron. 2023, 5, 1220387. [Google Scholar] [CrossRef]
- Arriagada, O.; Cacciuttolo, F.; Cabeza, R.A.; Carrasco, B.; Schwember, A.R. A Comprehensive Review on Chickpea (Cicer arietinum L.) Breeding for Abiotic Stress Tolerance and Climate Change Resilience. Int. J. Mol. Sci. 2022, 23, 6794. [Google Scholar] [CrossRef]
- Behera, S.K.; Shukla, A.K.; Tiwari, P.K.; Tripathi, A.; Singh, P.; Trivedi, V.; Patra, A.K.; Das, S. Classification of Pigeonpea (Cajanus cajan (L.) Millsp.) Genotypes for Zinc Efficiency. Plants 2020, 9, 952. [Google Scholar] [CrossRef]
- Kim, S.K.; Nair, R.M.; Lee, J.; Lee, S.-H. Genomic Resources in Mungbean for Future Breeding Programs. Front. Plant Sci. 2015, 6, 626. [Google Scholar] [CrossRef] [PubMed]
- Jha, U.C.; Shafi, S.; Tallury, S.; Nayyar, H.; Ciampitti, I.A.; Siddique, K.H.M.; Prasad, P.V.V. Differential Physiological and Yield Responses of Selected Mung Bean (Vigna radiata (L.) R. Wilczek) Genotypes to Various High-Temperature Stress Regimes. Sci. Rep. 2025, 15, 1034. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Seijo, G.; Freitas, F.O.; Valls, J.F.M.; Leal-Bertioli, S.C.M.; Moretzsohn, M.C. An Overview of Peanut and Its Wild Relatives. Plant Genet. Resour. 2011, 9, 134–149. [Google Scholar] [CrossRef]
- Liu, Y.; Shao, L.; Zhou, J.; Li, R.; Pandey, M.K.; Han, Y.; Cui, F.; Zhang, J.; Guo, F.; Chen, J.; et al. Genomic Insights into the Genetic Signatures of Selection and Seed Trait Loci in Cultivated Peanut. J. Adv. Res. 2022, 42, 237–248. [Google Scholar] [CrossRef]
- Guo, B.; Sun, L.; Jiang, S.; Ren, H.; Sun, R.; Wei, Z.; Hong, H.; Luan, X.; Wang, J.; Wang, X.; et al. Soybean Genetic Resources Contributing to Sustainable Protein Production. Theor. Appl. Genet. 2022, 135, 4095–4121. [Google Scholar] [CrossRef]
- Petereit, J.; Marsh, J.I.; Bayer, P.E.; Danilevicz, M.F.; Thomas, W.J.W.; Batley, J.; Edwards, D. Genetic and Genomic Resources for Soybean Breeding Research. Plants 2022, 11, 1181. [Google Scholar] [CrossRef]
- Zhuang, Y.; Li, X.; Hu, J.; Xu, R.; Zhang, D. Expanding the Gene Pool for Soybean Improvement with Its Wild Relatives. aBIOTECH 2022, 3, 115–125. [Google Scholar] [CrossRef]
- Tan, X.L.; Azam-Ali, S.; Von Goh, E.; Mustafa, M.; Chai, H.H.; Ho, W.K.; Mayes, S.; Mabhaudhi, T.; Azam-Ali, S.; Massawe, F. Bambara Groundnut: An Underutilized Leguminous Crop for Global Food Security and Nutrition. Front. Nutr. 2020, 7, 601496. [Google Scholar] [CrossRef]
- Tanzi, A.S.; Eagleton, G.E.; Ho, W.K.; Wong, Q.N.; Mayes, S.; Massawe, F. Winged Bean (Psophocarpus tetragonolobus (L.) DC.) for Food and Nutritional Security: Synthesis of Past Research and Future Direction. Planta 2019, 250, 911–931. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.K.; Singh, M.K. Horse Gram- an Underutilized Nutraceutical Pulse Crop: A Review. J. Food Sci. Technol. 2015, 52, 2489–2499. [Google Scholar] [CrossRef] [PubMed]
- George, T.T.; Obilana, A.O.; Oyeyinka, S.A. The Prospects of African Yam Bean: Past and Future Importance. Heliyon 2020, 6, e05458. [Google Scholar] [CrossRef]
- Odeku, O.A.; Ogunniyi, Q.A.; Ogbole, O.O.; Fettke, J. Forgotten Gems: Exploring the Untapped Benefits of Underutilized Legumes in Agriculture, Nutrition, and Environmental Sustainability. Plants 2024, 13, 1208. [Google Scholar] [CrossRef] [PubMed]
- Çakir, Ö.; Uçarli, C.; Tarhan, Ç.; Pekmez, M.; Turgut-Kara, N. Nutritional and Health Benefits of Legumes and Their Distinctive Genomic Properties. Food Sci. Tech. 2019, 39, 1–12. [Google Scholar] [CrossRef]
- Silver, W.L.; Perez, T.; Mayer, A.; Jones, A.R. The Role of Soil in the Contribution of Food and Feed. Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20200181. [Google Scholar] [CrossRef]
- Tan, Z.X.; Lal, R.; Wiebe, K.D. Global Soil Nutrient Depletion and Yield Reduction. J. Sustain. Agric. 2005, 26, 123–146. [Google Scholar] [CrossRef]
- Pires, D.; Orlando, V.; Collett, R.L.; Moreira, D.; Costa, S.R.; Inácio, M.L. Linking Nematode Communities and Soil Health under Climate Change. Sustainability 2023, 15, 11747. [Google Scholar] [CrossRef]
- Zhang, C.; Wright, I.J.; Nielsen, U.N.; Geisen, S.; Liu, M. Linking Nematodes and Ecosystem Function: A Trait-Based Framework. Trends Ecol. Evol. 2024, 39, 644–653. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From Saprophytes to Endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Zhang, X.; Harper, R.; Karsisto, M.; Lindstrom, K. Diversity of Rhizobium Bacteria Isolated from the Root Nodules of Leguminous Trees. Int. J. Syst. Bacteriol. 1991, 41, 104–113. [Google Scholar] [CrossRef]
- Lepetit, M.; Brouquisse, R. Control of the Rhizobium–Legume Symbiosis by the Plant Nitrogen Demand Is Tightly Integrated at the Whole Plant Level and Requires Inter-Organ Systemic Signaling. Front. Plant Sci. 2023, 14, 1114840. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.S.; Peoples, M.B.; Boddey, R.M.; Gresshoff, P.M.; Hauggaard-Nielsen, H.; Alves, B.J.R.; Morrison, M.J. Legumes for Mitigation of Climate Change and the Provision of Feedstock for Biofuels and Biorefineries. A Review. Agron. Sustain. Dev. 2012, 32, 329–364. [Google Scholar] [CrossRef]
- Wang, W.; Li, M.-Y.; Wen, Q.-H.; Ma, Y.; Zhang, Z.-M.; Rehman, M.M.U.; Mo, F.; Tao, H.-Y.; Ma, B.-L.; Whalen, J.K.; et al. Cereal-Legume Intercropping Stimulates Straw Decomposition and Promotes Soil Organic Carbon Stability. Sci. China Life Sci. 2025, 68, 1498–1508. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, S.; Fu, B.; Li, Z.; Lü, Y. Plantation Understorey Legume Functional Groups Enhance Soil Organic Carbon Sequestration by Promoting Species Richness. Land Degrad. Dev. 2023, 34, 2177–2188. [Google Scholar] [CrossRef]
- Wang, J.; Sun, P.; Li, Y.; Liu, Y.; Yu, J.; Ma, X.; Sun, S.; Yang, N.; Xia, R.; Lei, T.; et al. Hierarchically Aligning 10 Legume Genomes Establishes a Family-Level Genomics Platform. Plant Physiol. 2017, 174, 284–300. [Google Scholar] [CrossRef]
- Yuvaraj, M.; Pandiyan, M.; Gayathri, P. Role of Legumes in Improving Soil Fertility Status. In Legume Crops-Prospects, Production and Uses; Hasanuzzaman, M., Ed.; IntechOpen: London, UK, 2020. [Google Scholar]
- Ghosh, P.K.; Venkatesh, M.S.; Hazra, K.K.; Kumar, N. Long-Term Effect of Pulses and Nutrient Management on Soil Organic Carbon Dynamics and Sustainability on an Inceptisol of Indo-Gangetic Plains of India. Exp. Agric. 2012, 48, 473–487. [Google Scholar] [CrossRef]
- Stagnari, F.; Maggio, A.; Galieni, A.; Pisante, M. Multiple Benefits of Legumes for Agriculture Sustainability: An Overview. Chem. Biol. Technol. Agric. 2017, 4, 2. [Google Scholar] [CrossRef]
- Amthauer Gallardo, L.; Everwand, G.; Dauber, J. Effects of Legume Crops on Biodiversity. In Legumes Translated Report 2; European Union: Brussels, Belgium, 2022. [Google Scholar]
- Nelson, J.L.; Hunt, L.G.; Lewis, M.T.; Hamby, K.A.; Hooks, C.R.R.; Dively, G.P. Arthropod Communities in Warm and Cool Grass Riparian Buffers and Their Influence on Natural Enemies in Adjacent Crops. Agric. Ecosyst. Environ. 2018, 257, 81–91. [Google Scholar] [CrossRef]
- Everwand, G.; Cass, S.; Dauber, J.; Williams, M.; Stout, J. Legume Crops and Biodiversity. In Legumes in Cropping Systems; CABI: Wallingford, UK, 2017; pp. 55–69. [Google Scholar]
- Marzinzig, B.; Brünjes, L.; Biagioni, S.; Behling, H.; Link, W.; Westphal, C. Bee Pollinators of Faba Bean (Vicia faba L.) Differ in Their Foraging Behaviour and Pollination Efficiency. Agric. Ecosyst. Environ. 2018, 264, 24–33. [Google Scholar] [CrossRef]
- Carbas, B.; Machado, N.; Pathania, S.; Brites, C.; Rosa, E.A.; Barros, A.I. Potential of Legumes: Nutritional Value, Bioactive Properties, Innovative Food Products, and Application of Eco-Friendly Tools for Their Assessment. Food Rev. Int. 2023, 39, 160–188. [Google Scholar] [CrossRef]
- Maphosa, Y.; Jideani, V.A. The Role of Legumes in Human Nutrition. In Functional Food-Improve Health Through Adequate Food; Hueda, M.C., Ed.; IntechOpen: London, UK, 2017. [Google Scholar]
- Yanni, A.E.; Iakovidi, S.; Vasilikopoulou, E.; Karathanos, V.T. Legumes: A Vehicle for Transition to Sustainability. Nutrients 2023, 16, 98. [Google Scholar] [CrossRef]
- Multescu, M.; Culetu, A.; Susman, I.E. Screening of the Nutritional Properties, Bioactive Components, and Antioxidant Properties in Legumes. Foods 2024, 13, 3528. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Z.; Shen, A.; Zhang, T.; Jiang, L.; El-Seedi, H.; Zhang, G.; Sui, X. Legumes as an Alternative Protein Source in Plant-Based Foods: Applications, Challenges, and Strategies. Curr. Res. Food Sci. 2024, 9, 100876. [Google Scholar] [CrossRef]
- Trinidad, T.P.; Mallillin, A.C.; Loyola, A.S.; Sagum, R.S.; Encabo, R.R. The Potential Health Benefits of Legumes as a Good Source of Dietary Fibre. Br. J. Nutr. 2010, 103, 569–574. [Google Scholar] [CrossRef]
- Tiwari, U.; Cummins, E. Legume Fiber Characterization, Functionality, and Process Effects. In Pulse Foods; Elsevier: Amsterdam, The Netherlands, 2021; pp. 147–175. [Google Scholar]
- Ambika, S.R.D.S.; Kyada, A.D.; Ragi, S. Introduction to Legumes: Overview and Its Importance for Food Security. In Futuristic Trends in Agriculture and Allied Sciences; Bhatt, S.S., Patel, A., Kumar, S., Nautiyal, M., Eds.; Integrated Publishers: New Delhi, India, 2023; pp. 95–106. [Google Scholar]
- Martín-Cabrejas, M.A. Legumes: Nutritional Quality, Processing and Potential Health Benefits; Martín-Cabrejas, M.Á., Ed.; Royal Society of Chemistry: Cambridge, UK, 2019; ISBN 978-1-78801-161-7. [Google Scholar]
- Höhn, A.; Weber, D.; Jung, T.; Ott, C.; Hugo, M.; Kochlik, B.; Kehm, R.; König, J.; Grune, T.; Castro, J.P. Happily (n)Ever after: Aging in the Context of Oxidative Stress, Proteostasis Loss and Cellular Senescence. Redox Biol. 2017, 11, 482–501. [Google Scholar] [CrossRef]
- Samtiya, M.; Aluko, R.E.; Dhewa, T. Plant Food Anti-Nutritional Factors and Their Reduction Strategies: An Overview. Food Prod. Process. Nutr. 2020, 2, 6. [Google Scholar] [CrossRef]
- Messina, M. Impact of Soy Foods on the Development of Breast Cancer and the Prognosis of Breast Cancer Patients. Complement. Med. Res. 2016, 23, 75–80. [Google Scholar] [CrossRef]
- Alcázar-Valle, M.; Lugo-Cervantes, E.; Mojica, L.; Morales-Hernández, N.; Reyes-Ramírez, H.; Enríquez-Vara, J.N.; García-Morales, S. Bioactive Compounds, Antioxidant Activity, and Antinutritional Content of Legumes: A Comparison between Four Phaseolus Species. Molecules 2020, 25, 3528. [Google Scholar] [CrossRef]
- Bautista-Expósito, S.; Vandenberg, A.; Peñas, E.; Frias, J.; Martínez-Villaluenga, C. Lentil and Fava Bean With Contrasting Germination Kinetics: A Focus on Digestion of Proteins and Bioactivity of Resistant Peptides. Front. Plant Sci. 2021, 12, 754287. [Google Scholar] [CrossRef] [PubMed]
- Salaria, S.; Boatwright, J.L.; Thavarajah, P.; Kumar, S.; Thavarajah, D. Protein Biofortification in Lentils (Lens culinaris Medik.) Toward Human Health. Front. Plant Sci. 2022, 13, 869713. [Google Scholar] [CrossRef] [PubMed]
- Merga, B.; Haji, J. Economic Importance of Chickpea: Production, Value, and World Trade. Cogent Food Agric. 2019, 5, 1615718. [Google Scholar] [CrossRef]
- Shaikh, M.; Sunooj, K.V.; Rahman, M.H.; Navaf, M.; Ali, T.M. Lentils: A Recent Review on Global Trade and Popular Regional Cuisines. Legume Sci. 2024, 6, e252. [Google Scholar] [CrossRef]
- Xavier, B. Future Use Prospects of Legumes through Improvement and the Challenges Faced. In Production and Utilization of Legumes-Progress and Prospects; IntechOpen: London, UK, 2023. [Google Scholar]
- Vadez, V.; Berger, J.D.; Warkentin, T.; Asseng, S.; Ratnakumar, P.; Rao, K.P.C.; Gaur, P.M.; Munier-Jolain, N.; Larmure, A.; Voisin, A.-S.; et al. Adaptation of Grain Legumes to Climate Change: A Review. Agron. Sustain. Dev. 2012, 32, 31–44. [Google Scholar] [CrossRef]
- van Zanten, H.H.E.; Bikker, P.; Mollenhorst, H.; Meerburg, B.G.; de Boer, I.J.M. Environmental Impact of Replacing Soybean Meal with Rapeseed Meal in Diets of Finishing Pigs. Animal 2015, 9, 1866–1874. [Google Scholar] [CrossRef]
- Piquet-Pissaloux, A. Environmental Footprints of Legumes-Based Agroecosystems for Sustainable Development. In Advances in Legumes for Sustainable Intensification; Meena, R.S., Kumar, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 421–440. [Google Scholar]
- Liu, K.; Machado, P.V.F.; Lin, S.; Drury, C.F.; Lemke, R.L. Soil Nitrous Oxide Emissions from Wheat-Based Rotations with Different Types of Pulse Crops. J. Environ. Manag. 2024, 370, 122830. [Google Scholar] [CrossRef]
- Pande, S.; Sharma, M.; Kumari, S.; Gaur, P.; Chen, W.; Kaur, L.; MacLeod, W.; Basandrai, A.; Basandrai, D.; Bakr, A.; et al. Integrated Foliar Diseases Management of Legumes. In Proceedings of the International Conference on Grain Legumes: Quality Improvement, Value Addition and Trade, Kanpur, India, 14–19 February 2009; pp. 143–147. [Google Scholar]
- Rubiales, D.; Fondevilla, S.; Chen, W.; Gentzbittel, L.; Higgins, T.J.V.; Castillejo, M.A.; Singh, K.B.; Rispail, N. Achievements and Challenges in Legume Breeding for Pest and Disease Resistance. CRC Crit. Rev. Plant Sci. 2015, 34, 195–236. [Google Scholar] [CrossRef]
- Hull, R. Alfalfa Mosaic Virus. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 1969; pp. 365–433. [Google Scholar]
- Bagewadi, B.; Fauquet, C.M. Plant Virus Control by Post-Transcriptional Gene Silencing. In Encyclopedia of Agriculture and Food Systems; Van Alfen, N.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 472–488. [Google Scholar]
- Garrido-Ramirez, E.R.; Sudarshana, M.R.; Gilbertson, R.L. Bean Golden Yellow Mosaic Virus from Chiapas, Mexico: Characterization, Pseudorecombination with Other Bean-Infecting Geminiviruses and Germ Plasm Screening. Phytopathology 2000, 90, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Roy, B.; Nekkanti, A.; Das, A.; Dhar, S.; Mukherjee, S.K. Transovarial Transmission of Dolichos Yellow Mosaic Virus by Its Vector, Bemisia Tabaci Asia II 1. Front. Microbiol. 2021, 12, 755155. [Google Scholar] [CrossRef] [PubMed]
- Akram, M.; Singh, N.P. Yellow Mosaic of Mungbean and Urdbean: Current Status and Future Strategies. J. Food Legumes 2016, 29, 77–93. [Google Scholar]
- Fontenele, R.S.; Poppiel, R.; Matos, V.O.R.L.; Costa, F.; Faria, J.C.; Ribeiro, S.G. First Report of Macroptilium Yellow Spot Virus in Desmodium Glabrum in Brazil. Plant Dis. 2016, 100, 657. [Google Scholar] [CrossRef]
- Mishra, G.P.; Dikshit, H.K.; Ramesh, S.V.; Tripathi, K.; Kumar, R.R.; Aski, M.; Singh, A.; Roy, A.; Priti; Kumari, N.; et al. Yellow Mosaic Disease (YMD) of Mungbean (Vigna radiata (L.) Wilczek): Current Status and Management Opportunities. Front. Plant Sci. 2020, 11, 918. [Google Scholar] [CrossRef]
- Kumar, S.; Tanti, B.; Mukherjee, S.K.; Sahoo, L. Molecular Characterization and Infectivity of Mungbean Yellow Mosaic India Virus Associated with Yellow Mosaic Disease of Cowpea and Mungbean. Biocatal. Agric. Biotechnol. 2017, 11, 183–191. [Google Scholar] [CrossRef]
- Chakraborty, S. Tomato Leaf Curl Viruses from India. In Encyclopedia of Virology; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Elsevier: Amsterdam, The Netherlands, 2008; pp. 124–133. [Google Scholar]
- Lapidot, M.; Polston, J.E. Resistance to Tomato Yellow Leaf Curl Virus in Tomato. In Natural Resistance Mechanisms of Plants to Viruses; Loebenstein, G., Carr, J.P., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 503–520. [Google Scholar]
- Brunt, A.A.; Kenten, R.H. Cowpea Mild Mottle, a Newly Recognized Virus Infecting Cowpeas (Vigna unguiculata) in Ghana. Ann. Appl. Biol. 1973, 74, 67–74. [Google Scholar] [CrossRef]
- Fulton, J.; Cumberland, D.; Hodgsen, O.; Amsoy, C.; Vickery, W. Bean Pod Mottle Virus: Occurrence in Nebraska and Seed Transmission in Soybeans. Plant Dis. 1983, 67, 230–233. [Google Scholar] [CrossRef]
- Booker, H.M.; Umaharan, P.; McDavid, C.R. Effect of Cowpea Severe Mosaic Virus on Crop Growth Characteristics and Yield of Cowpea. Plant Dis. 2005, 89, 515–520. [Google Scholar] [CrossRef][Green Version]
- Abdullahi, I.; Ikotun, T.; Winter, S.; Thottappilly, G.; Atiri, G.I. Investigation on Seed Transmission of Cucumber Mosaic Virus in Cowpea. Afr. Crop Sci. J. 2001, 9, 677–684. [Google Scholar] [CrossRef]
- Hill, J.H.; Whitham, S.A. Control of Virus Diseases in Soybeans. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2014; pp. 355–390. [Google Scholar]
- Lamprecht, R.L.; Kasdorf, G.G.F.; Stiller, M.; Staples, S.M.; Nel, L.H.; Pietersen, G. Soybean Blotchy Mosaic Virus, a New Cytorhabdovirus Found in South Africa. Plant Dis. 2010, 94, 1348–1354. [Google Scholar] [CrossRef][Green Version]
- Patil, B.L.; Kumar, P.L. Pigeonpea Sterility Mosaic Virus: A Legume-infecting Emaravirus from South Asia. Mol. Plant Pathol. 2015, 16, 775–786. [Google Scholar] [CrossRef]
- Sandra, N.; Tripathi, A.; Dikshit, H.K.; Mandal, B.; Jain, R.K. Seed Transmission of a Distinct Soybean Yellow Mottle Mosaic Virus Strain Identified from India in Natural and Experimental Hosts. Virus Res. 2020, 280, 197903. [Google Scholar] [CrossRef]
- Reddy, A.S.; Rao, R.D.V.J.P.; Thirumala-Devi, K.; Reddy, S.V.; Mayo, M.A.; Roberts, I.; Satyanarayana, T.; Subramaniam, K.; Reddy, D.V.R. Occurrence of Tobacco Streak Virus on Peanut (Arachis hypogaea) in India. Plant Dis. 2002, 86, 173–178. [Google Scholar] [CrossRef]
- Tamada, T. Aphid Transmission and Host Range of Soybean Dwarf Virus. Jpn. J. Phytopathol. 1970, 36, 266–274. [Google Scholar] [CrossRef]
- Lotfipour, M.; Behjatnia, S.A.A.; Dall’Ara, M.; Ratti, C. The Full-Length Genome Characterization and Diversity of Faba Bean Necrotic Stunt Virus in Iran. Eur. J. Plant Pathol. 2020, 157, 239–250. [Google Scholar] [CrossRef]
- Zhang, C.; Zheng, H.; Yan, D.; Han, K.; Song, X.; Liu, Y.; Zhang, D.; Chen, J.; Yan, F. Complete Genomic Characterization of Milk Vetch Dwarf Virus Isolates from Cowpea and Broad Bean in Anhui Province, China. Arch. Virol. 2017, 162, 2437–2440. [Google Scholar] [CrossRef] [PubMed]
- Sharman, M.; Thomas, J.E.; Tree, D.; Persley, D.M. Natural Host Range and Thrips Transmission of Capsicum Chlorosis Virus in Australia. Australas. Plant Pathol. 2020, 49, 45–51. [Google Scholar] [CrossRef]
- Lakshmi, K.V.; Wightman, J.A.; Reddy, D.V.R.; Rao, G.V.R.; Buiel, A.A.M.; Reddy, D.D.R. Transmission of Peanut Bud Necrosis Virus by Thrips Palmi in India. In Thrips Biology and Management; Parker, B.L., Skinner, M., Lewis, T., Eds.; Springer: Boston, MA, USA, 1995; pp. 179–184. [Google Scholar]
- Camelo-García, V.M.; Lima, E.F.B.; Mansilla-Córdova, P.J.; Rezende, J.A.M.; Kitajima, E.W.; Barreto, M. Occurrence of Groundnut Ringspot Virus on Brazilian Peanut Crops. J. Gen. Plant Pathol. 2014, 80, 282–286. [Google Scholar] [CrossRef]
- Groves, C.; German, T.; Dasgupta, R.; Mueller, D.; Smith, D.L. Seed Transmission of Soybean Vein Necrosis Virus: The First Tospovirus Implicated in Seed Transmission. PLoS ONE 2016, 11, e0147342. [Google Scholar] [CrossRef]
- Allen, W.R.; Broadbent, A.B. Transmission of Tomato Spotted Wilt Virus in Ontario Greenhouses by Frankliniella Occidentalis. Can. J. Plant Pathol. 1986, 8, 33–38. [Google Scholar] [CrossRef]
- Worrall, E.A.; Wamonje, F.O.; Mukeshimana, G.; Harvey, J.J.W.; Carr, J.P.; Mitter, N. Bean Common Mosaic Virus and Bean Common Mosaic Necrosis Virus: Relationships, Biology, and Prospects for Control. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2015; Volume 93, pp. 1–46. [Google Scholar]
- Harrison, B.D.; Muniyappa, V.; Swanson, M.M.; Roberts, I.M.; Robinson, D.J. Recognition and Differentiation of Seven Whitefly-transmitted Geminiviruses from India, and Their Relationships to African Cassava Mosaic and Thailand Mung Bean Yellow Mosaic Viruses. Ann. Appl. Biol. 1991, 118, 299–308. [Google Scholar] [CrossRef]
- Damiri, B.V.; Al-Shahwan, I.M.; Al-Saleh, M.A.; Abdalla, O.A.; Amer, M.A. Identification and Characterization of Cowpea Aphid-Borne Mosaic Virus Isolates in Saudi Arabia. J. Plant Pathol. 2013, 95, 79–85. [Google Scholar]
- Adams, D.B.; Kuhn, C.W. Seed Transmission of Peanut Mottle Virus in Peanuts. Phytopathology 1977, 77, 1126–1129. [Google Scholar] [CrossRef]
- Rehman, F.U.; Kalsoom, M.; Adnan, M.; Naz, N.; Ahmad Nasir, T.; Ali, H.; Shafique, T.; Murtaza, G.; Anwar, S.; Arshad, M.A. Soybean Mosaic Disease (SMD): A Review. Egypt. J. Basic Appl. Sci. 2021, 8, 12–16. [Google Scholar] [CrossRef]
- Uyemoto, J.K.; Grogan, R.G. Southern Bean Mosaic Virus: Evidence for Seed Transmission in Bean Embryos. Phytopathology 1977, 77, 1190–1196. [Google Scholar] [CrossRef]
- Wei, Z.Y.; Jiang, C.; Mao, C.Y.; Zhang, H.H.; Miao, R.P.; Chen, J.P.; Sun, Z.T. Occurrence of Soybean Yellow Common Mosaic Virus in Soybean in China Showing Yellow Common Mosaic Disease. Plant Dis. 2021, 105, 1236. [Google Scholar] [CrossRef]
- Waliyar, F.; Kumar, L.; Ntare, B.R.; Monyo, E.; Nigam, S.; Reddy, A.; Osiru, M.; Diallo, A.T. A Century of Research on Groundnut Rosette Disease and Its Management. Information Bulletin No. 75; ICRISAT: Hyderabad, India, 2007. [Google Scholar]
- Hull, R.; Adams, A.N. Groundnut Rosette and Its Assistor Virus. Ann. Appl. Biol. 1968, 62, 139–145. [Google Scholar] [CrossRef]
- France, R.A.; Abawi, G.S. Interaction between Meloidogyne Incognita and Fusarium oxysporum f. sp. Phaseoli on Selected Bean Genotypes. J. Nematol. 1994, 26, 467–474. [Google Scholar]
- Kumar, R.; Ahmad, S.; Saxena, S.K. Disease Complex in Chickpea Involving Meloidogyne Incognita and Fusarium oxysporum. Int. Nematol. Netw. Newsl. 1988, 5, 12–14. [Google Scholar]
- Maheshwari, T.U.; Sharma, S.B.; Reddy, D.D.; Haware, M.P. Co-Infection of Wilt-Resistant Chickpeas by Fusarium oxysporum f. sp. Ciceri and Meloidogyne Javanica. J. Nematol. 1995, 27, 649–653. [Google Scholar]
- De, R.K.; Ali, S.S.; Dwivedi, R.P. Effect of Interaction between Fusarium oxysporum f. sp. Lentis and Meloidogyne Javanica on Lentil. Indian J. Pulses Res. 2001, 14, 71–73. [Google Scholar]
- Abdel-Momen, S.M.; Starr, J.L. Meloidogyne Javanica-Rhizoctonia Solani Disease Complex of Peanut. Fundam. Appl. Nematol. 1998, 21, 611–618. [Google Scholar]
- Siddiqui, Z.A.; Mahmood, I. Effects of Meloidogyne Incognita, Fusarium oxysporum f. sp. Pisi, Rhizobium sp. and Different Soil Types on Growth, Chlorophyll, and Carotenoid Pigments of Pea. Isr. J. Plant Sci. 1999, 47, 251–256. [Google Scholar] [CrossRef]
- Vats, R.; Dalal, M.R. Interaction between Rotylenchulus Reniformis and Fusarium oxysporum f. sp. Pisi on Pea (Pisum sativum L.). Ann. Biol. 1997, 13, 239–242. [Google Scholar]
- Rupe, J.C. Frequency and Pathogenicity of Fusarium Solani Recovered from Soybeans with Sudden Death Syndrome. Plant Dis. 1989, 73, 581–584. [Google Scholar] [CrossRef]
- Kaitany, R.; Melakeberhan, H.; Bird, G.W.; Safir, G. Association of Phytophthora Sojae with Heterodera Glycines and Nutrient-Stressed Soybeans. Nematropica 2000, 30, 193–199. [Google Scholar]
- Schwartz, H.F.; Steadman, J.R.; Hall, R.; Forster, R.L. Compendium of Bean Diseases; APS Press: Saint Paul, MN, USA, 2005. [Google Scholar]
- Roberts, S.J.; Phelps, K.; McKeown, B.M.; Heath, M.C.; Cockerell, V. Effect of Pea Bacterial Blight (Pseudomonas syringae pv. pisi) on the Yield of Spring Sown Combining Peas (Pisum sativum). Ann. Appl. Biol. 1995, 126, 61–73. [Google Scholar] [CrossRef]
- Elphinstone, J.G. The Current Bacterial Wilt Situation: A Global Overview. In Bacterial Wilt Disease and the Ralstonia Solanacearum Species Complex; Allen, C., Prior, P., Hayward, A.C., Eds.; APS Press: Saint Paul, MN, USA, 2005; pp. 9–28. [Google Scholar]
- Cohen, Y.; Coffey, M.D. Fusarium Wilt in Beans. Phytopathology 2003, 93, 408–417. [Google Scholar]
- Kumar, A.; Pathak, H.; Bhadauria, S.; Sudan, J. Aflatoxin Contamination in Food Crops: Causes, Detection, and Management: A Review. Food Prod. Process. Nutr. 2021, 3, 17. [Google Scholar] [CrossRef]
- Giachero, M.L.; Declerck, S.; Marquez, N. Phytophthora Root Rot: Importance of the Disease, Current and Novel Methods of Control. Agronomy 2022, 12, 610. [Google Scholar] [CrossRef]
- Gaulin, E.; Jacquet, C.; Bottin, A.; Dumas, B. Root Rot Disease of Legumes Caused by Aphanomyces Euteiches. Mol. Plant Pathol. 2007, 8, 539–548. [Google Scholar] [CrossRef]
- Chang, K.F.; Hwang, S.F.; Ahmed, H.U.; Strelkov, S.E.; Conner, R.L.; Gossen, B.D.; Bing, D.J.; Turnbull, G.D. Yield Loss and Management of Downy Mildew on Field Pea in Alberta, Canada. Crop Prot. 2013, 46, 23–28. [Google Scholar] [CrossRef]
- CABI. Peronospora Manshurica (Soybean Downy Mildew); CABI Compendium: Wallingford, UK, 2021. [Google Scholar]
- Manjunatha, N.; Rangaswamy, K.T.; Nagaraju, N.; Reddy, M.K.; Prameela, H.A.; Manjunath, S.H. Biological Relationship of Bean Common Mosaic Virus (BCMV) Infecting Cowpea with Leguminous Plant Species. J. Appl. Nat. Sci. 2017, 9, 2170–2174. [Google Scholar] [CrossRef]
- Parameswari, B.; Bajaru, D.; Karthikaiselvi, N.; Sivaraj, P.; Pranusha, P.; Brahmi, P.; Mangrauthia, S.; Saravanan, L.; Chalam, V.; Kodaru, A. Interception of Bean Common Mosaic Virus in Bambara Groundnut Accessions Imported from Ghana through RT-PCR. Indian. J. Plant Prot. 2022, 50, 80–85. [Google Scholar]
- Tang, M.; Feng, X. Bean Common Mosaic Disease: Etiology, Resistance Resource, and Future Prospects. Agronomy 2022, 13, 58. [Google Scholar] [CrossRef]
- Rahman, M.M.; Jarugula, S.; Bagewadi, B.; Fayad, A.; Karasev, A.V.; Naidu, R.A. Characterization of a New, Country Bean (Lablab purpureus) Lineage of Bean Common Mosaic Necrosis Virus. Plant Dis. 2024, 108, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, A.; Shobhana, V.G.; Sudha, M.; Raveendran, M.; Senthil, N.; Pandiyan, M.; Nagarajan, P. Mungbean Yellow Mosaic Virus (MYMV): A Threat to Green Gram (Vigna radiata) Production in Asia. Int. J. Pest. Manag. 2014, 60, 314–324. [Google Scholar] [CrossRef]
- Shahid, M.S.; Ikegami, M.; Natsuaki, K.T. First Report of Mungbean Yellow Mosaic India Virus on Lima Bean Affected by Yellow Mosaic Disease in Nepal. Australas. Plant Dis. Notes 2012, 7, 85–89. [Google Scholar] [CrossRef]
- Singh, S.K.; Chakraborty, S.; Singh, A.K.; Pandey, P.K. Cloning, Restriction Mapping and Phylogenetic Relationship of Genomic Components of MYMIV from Lablab purpureus. Bioresour. Technol. 2006, 97, 1807–1814. [Google Scholar] [CrossRef]
- Rienzie, R.; De Costa, D.; Wickramaarachchi, T. Transmission and Host Range of Horsegram Yellow Mosaic Virus (HgYMV) Causing Common Bean (Phaseolus vulgaris L.) Yellowing Disease in Sri Lanka. J. Natl. Sci. Found. 2020, 48, 81. [Google Scholar] [CrossRef]
- Zanardo, L.G.; Carvalho, C.M. Cowpea Mild Mottle Virus (Carlavirus, Betaflexiviridae): A Review. Trop. Plant Pathol. 2017, 42, 417–430. [Google Scholar] [CrossRef]
- Medeiros, L.d.S.A.; de Oliveira, I.A.; Kitajima, E.W.; Eiras, M.; Pereira, H.J.; Ribeiro, S.G.; Matos, K.d.S.; Beserra Júnior, J.E.A. A Survey of RNA Genome Viruses in Lima Bean Crops of Northeastern Brazil. Bragantia 2020, 79, 407–416. [Google Scholar] [CrossRef]
- Das, S. Characterization of Viruses in Legume Vegetables and Identification of Aphid Resistance in Lentil Germplasm; Washington State University: Pullman, WA, USA, 2021. [Google Scholar]
- Bashir, M.; Ahmad, Z.; Ghafoor, A. Cowpea Aphid-Borne Mosaic Potyvirus: A Review. Int. J. Pest. Manag. 2002, 48, 155–168. [Google Scholar] [CrossRef]
- Karthikeyan, G.; Manoranjitham, S.K. Ground Nut (Peanut). In Viral Diseases of Field and Horticultural Crops; Awasthi, L.P., Ed.; Elsevier: Amsterdam, The Netherlands, 2024; pp. 163–170. [Google Scholar]
- Jain, R.K.; Sudeep, B.; Ramiah, M. Natural Infection of Groundnut Bud Necrosis Virus in Cotton and Sem. Indian J. Virol. 2005, 16, 50. [Google Scholar]
- Sastry, K.S.; Mandal, B.; Hammond, J.; Scott, S.W.; Briddon, R.W. Encyclopedia of Plant Viruses and Viroids; Springer: New Delhi, India, 2019; ISBN 978-81-322-3911-6. [Google Scholar]
- Nam, M.; Kim, S.M.; Domier, L.L.; Koh, S.; Moon, J.K.; Choi, H.S.; Kim, H.G.; Moon, J.S.; Lee, S.-H. Nucleotide Sequence and Genomic Organization of a Newly Identified Member of the Genus Carmovirus, Soybean Yellow Mottle Mosaic Virus, from Soybean. Arch. Virol. 2009, 154, 1679–1684. [Google Scholar] [CrossRef]
- Sandra, N.; Kumar, A.; Sharma, P.; Kapoor, R.; Jain, R.K.; Mandal, B. Diagnosis of a New Variant of Soybean Yellow Mottle Mosaic Virus with Extended Host-Range in India. Virusdisease 2015, 26, 304–314. [Google Scholar] [CrossRef]
- Beserra, A., Jr.; Evando, J.; Miguel Teixeira, J.W.; Marques Lima, K.J.; Eiras, M. Preliminary Survey of RNA Genome Viruses in Lima Bean; USDA-ARS/UNL Faculty: Lincoln, NE, USA, 2017. [Google Scholar]
- Kannan, R. Tobacco Streak Virus in Plants-A Review. Agric. Rev. 2012, 33, 333–340. [Google Scholar]
- Khan, M.S.; Tiwari, A.K.; Khan, A.A.; Ji, S.H.; Chun, S.C. Current Scenario of Tomato Yellow Leaf Curl Virus (TYLCV) and Its Possible Management: A Review. Vegetos-An. Int. J. Plant Res. 2013, 26, 139. [Google Scholar] [CrossRef]
- Sharma, N.; Prasad, M. An Insight into Plant–Tomato Leaf Curl New Delhi Virus Interaction. Nucleus 2017, 60, 335–348. [Google Scholar] [CrossRef]
- Hasan, M.; Akter, T.; Sano, Y. Identification of Milk Vetch Dwarf Virus from Mungbean (Vigna radiata L.) in Bangladesh. Indian Phytopathol. 2024, 77, 227–231. [Google Scholar] [CrossRef]
- Liu, J.-Z.; Fang, Y.; Pang, H. The Current Status of the Soybean-Soybean Mosaic Virus (SMV) Pathosystem. Front. Microbiol. 2016, 7, 1906. [Google Scholar] [CrossRef]
- Robinson, D.J.; Ryabov, E.V.; Raj, S.K.; Roberts, I.M.; Taliansky, M.E. Satellite RNA Is Essential for Encapsidation of Groundnut Rosette Umbravirus RNA by Groundnut Rosette Assistor Luteovirus Coat Protein. Virology 1999, 254, 105–114. [Google Scholar] [CrossRef]
- Culbreath, A.K.; Todd, J.W.; Brown, S.L. Epidemiology and Management of Tomato Spotted Wilt in Peanut. Annu. Rev. Phytopathol. 2003, 41, 53–75. [Google Scholar] [CrossRef]
- Demski, J.W. Source and Spread of Peanut Mottle Virus in Soybean and Peanut. Phytopathology 1975, 65, 917. [Google Scholar] [CrossRef]
- Nam, M.; Park, S.-J.; Kim, Y.-J.; Kim, J.-S.; Park, C.-Y.; Lee, J.-S.; Choi, H.-S.; Kim, J.-S.; Kim, H.-G.; Lee, S.-H. First Report of Peanut Stunt Virus on Glycine max in Korea. Plant Pathol. J. 2012, 28, 330. [Google Scholar] [CrossRef]
- Fontes, M.G.; Andrews da Silva, G.F.; Lima, M.F.; Fonseca, M.E.N.; Costa, A.F.; Silva-Filho, J.G.; Boiteux, L.S. First Report of Groundnut Ringspot Orthotospovirus Infecting Soybeans in Brazil. Plant Dis. 2019, 103, 777. [Google Scholar] [CrossRef]
- Bragard, C.; Baptista, P.; Chatzivassiliou, E.; Gonthier, P.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P.; Navas-Cortes, J.A.; et al. Pest Categorisation of Capsicum Chlorosis Virus. EFSA J. 2022, 20, e07337. [Google Scholar] [CrossRef] [PubMed]
- Chatzivassiliou, E.K. An Annotated List of Legume-Infecting Viruses in the Light of Metagenomics. Plants 2021, 10, 1413. [Google Scholar] [CrossRef]
- de Freitas-Vanzo, A.T.; Silva, C.d.C.d.; Chaves, V.C.A.; Garcia, M.H.; de Aquino, L.T.; Molina, R.d.O. Detection of Bean Golden Mosaic Virus in Fabaceae Family Plants. Braz. J. Anim. Environ. Res. 2021, 4, 1021–1032. [Google Scholar] [CrossRef]
- Sobrinho, R.R.; Xavier, C.A.D.; Pereira, H.M.d.B.; Lima, G.S.d.A.; Assunção, I.P.; Mizubuti, E.S.G.; Duffy, S.; Zerbini, F.M. Contrasting Genetic Structure between Two Begomoviruses Infecting the Same Leguminous Hosts. J. Gen. Virol. 2014, 95, 2540–2552. [Google Scholar] [CrossRef]
- Jones, A.T.; Kumar, P.L.; Saxena, K.B.; Kulkarni, N.K.; Muniyappa, V.; Waliyar, F. Sterility Mosaic Disease—The “Green Plague” of Pigeonpea: Advances in Understanding the Etiology, Transmission and Control of a Major Virus Disease. Plant Dis. 2004, 88, 436–445. [Google Scholar] [CrossRef]
- Maruthi, M.N.; Manjunatha, B.; Rekha, A.R.; Govindappa, M.R.; Colvin, J.; Muniyappa, V. Dolichos Yellow Mosaic Virus Belongs to a Distinct Lineage of Old World Begomoviruses; Its Biological and Molecular Properties. Ann. Appl. Biol. 2006, 149, 187–195. [Google Scholar] [CrossRef]
- Nicol, J.M.; Turner, S.J.; Coyne, D.L.; den Nijs, L.; Hockland, S.; Maafi, Z.T. Current Nematode Threats to World Agriculture. In Genomics and Molecular Genetics of Plant-Nematode Interactions; Jones, J.T., Gheysen, G., Fenoll, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 21–43. [Google Scholar]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 Plant-Parasitic Nematodes in Molecular Plant Pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Topalović, O.; Geisen, S. Nematodes as Suppressors and Facilitators of Plant Performance. New Phytol. 2023, 238, 2305–2312. [Google Scholar] [CrossRef] [PubMed]
- Parrado, L.M.; Quintanilla, M. Plant-Parasitic Nematode Disease Complexes as Overlooked Challenges to Crop Production. Front. Plant Sci. 2024, 15, 1439951. [Google Scholar] [CrossRef] [PubMed]
- Ratu, S.T.N.; Teulet, A.; Miwa, H.; Masuda, S.; Nguyen, H.P.; Yasuda, M.; Sato, S.; Kaneko, T.; Hayashi, M.; Giraud, E.; et al. Rhizobia Use a Pathogenic-like Effector to Hijack Leguminous Nodulation Signalling. Sci. Rep. 2021, 11, 2034. [Google Scholar] [CrossRef]
- Zhang, W.; Luo, X.; Mei, Y.; Yang, Q.; Zhang, A.; Chen, M.; Mei, Y.; Ma, C.; Du, Y.; Li, M.; et al. Priming of Rhizobial Nodulation Signaling in the Mycosphere Accelerates Nodulation of Legume Hosts. New Phytol. 2022, 235, 1212–1230. [Google Scholar] [CrossRef]
- Li, X.; Xiao, R. Molecular Dialogue in Legume-Rhizobium Symbiosis: Signaling Mechanisms and Genetic Insights. Rhizosphere 2025, 33, 101034. [Google Scholar] [CrossRef]
- Primieri, S.; Magnoli, S.M.; Koffel, T.; Stürmer, S.L.; Bever, J.D. Perennial, but Not Annual Legumes Synergistically Benefit from Infection with Arbuscular Mycorrhizal Fungi and Rhizobia: A Meta-analysis. New Phytol. 2022, 233, 505–514. [Google Scholar] [CrossRef]
- Liu, A.; Ku, Y.-S.; Contador, C.A.; Lam, H.-M. The Impacts of Domestication and Agricultural Practices on Legume Nutrient Acquisition through Symbiosis with Rrhizobia and Arbuscular Mycorrhizal Fungi. Front. Genet. 2020, 11, 583954. [Google Scholar] [CrossRef]
- Duan, H.-X.; Luo, C.-L.; Wang, X.; Cheng, Y.-S.; Abrar, M.; Batool, A. Responses of Legumes to Rhizobia and Arbuscular Mycorrhizal Fungi Under Abiotic Stresses: A Global Meta-Analysis. Agronomy 2024, 14, 2597. [Google Scholar] [CrossRef]
- Siddiqui, Z.A.; Mahmood, I. Role of Plant Symbionts in Nematode Management: A Review. Bioresour. Technol. 1995, 54, 217–226. [Google Scholar] [CrossRef]
- Gough, E.C.; Owen, K.J.; Zwart, R.S.; Thompson, J.P. Arbuscular Mycorrhizal Fungi Acted Synergistically with Bradyrhizobium sp. to Improve Nodulation, Nitrogen Fixation, Plant Growth and Seed Yield of Mung Bean (Vigna radiata) but Increased the Population Density of the Root-Lesion Nematode Pratylenchus Thornei. Plant Soil. 2021, 465, 431–452. [Google Scholar] [CrossRef]
- Elhady, A.; Hallmann, J.; Heuer, H. Symbiosis of Soybean with Nitrogen Fixing Bacteria Affected by Root Lesion Nematodes in a Density-Dependent Manner. Sci. Rep. 2020, 10, 1619. [Google Scholar] [CrossRef]
- Hussey, R.S. Host-Parasite Relationships and Associated Physiological Changes. In An Advanced Treatise on Meloidogyne Volume 1: Biology and Control; North Carolina State University Graphics: Raleigh, NC, USA, 1985; pp. 143–153. [Google Scholar]
- Wood, C.W.; Pilkington, B.L.; Vaidya, P.; Biel, C.; Stinchcombe, J.R. Genetic Conflict with a Parasitic Nematode Disrupts the Legume–Rhizobia Mutualism. Evol. Lett. 2018, 2, 233–245. [Google Scholar] [CrossRef]
- Costa, S.R.; Ng, J.L.P.; Mathesius, U. Interaction of Symbiotic Rhizobia and Parasitic Root-Knot Nematodes in Legume Roots: From Molecular Regulation to Field Application. Mol. Plant-Microbe Interact. 2021, 34, 470–490. [Google Scholar] [CrossRef]
- Barker, K.R. Antagonistic Interaction between Heterodera Glycines and Rhizobium Japonicum on Soybean. Phytopathology 1972, 62, 1201. [Google Scholar] [CrossRef]
- McGinnity, P.J.; Kapusta, G.; Myers, O. Soybean Cyst Nematode and Rhizobium Strain Influences on Soybean Nodulation and N2-fixation. Agron. J. 1980, 72, 785–789. [Google Scholar] [CrossRef]
- Mergaert, P.; Giraud, E. Pathogenic Nematodes Exploit Achilles’ Heel of Plant Symbioses. Trends Parasitol. 2024, 40, 873–875. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Zhu, X.; Liu, R.; Xiang, P.; Chen, J.; Liu, X.; Duan, Y.; Chen, L. Management of the Soybean Cyst Nematode Heterodera Glycines with Combinations of Different Rhizobacterial Strains on Soybean. PLoS ONE 2017, 12, e0182654. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Sikandar, A.; Zhao, Y.S.; Zhao, J.; Liu, D.; Zhu, X.F.; Liu, X.Y.; Fan, H.Y.; Chen, L.J.; Duan, Y.X. Effect of Culture Filtrate of Sinorhizobium Frediisneb183 on the Activity and Behavior of Soybean Cyst Nematode (Heterodera Glycines Ichinohe, 1952). Appl. Ecol. Environ. Res. 2020, 18, 1129–1140. [Google Scholar] [CrossRef]
- Danchin, E.G.J.; Guzeeva, E.A.; Mantelin, S.; Berepiki, A.; Jones, J.T. Horizontal Gene Transfer from Bacteria Has Enabled the Plant-Parasitic Nematode Globodera Pallida to Feed on Host-Derived Sucrose. Mol. Biol. Evol. 2016, 33, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Danchin, E.G.J.; Rosso, M.-N.; Vieira, P.; de Almeida-Engler, J.; Coutinho, P.M.; Henrissat, B.; Abad, P. Multiple Lateral Gene Transfers and Duplications Have Promoted Plant Parasitism Ability in Nematodes. Proc. Natl. Acad. Sci. USA 2010, 107, 17651–17656. [Google Scholar] [CrossRef] [PubMed]
- Goverse, A.; Smant, G. The Activation and Suppression of Plant Innate Immunity by Parasitic Nematodes. Annu. Rev. Phytopathol. 2014, 52, 243–265. [Google Scholar] [CrossRef]
- Koltai, H.; Dhandaydham, M.; Opperman, C.; Thomas, J.; Bird, D. Overlapping Plant Signal Transduction Pathways Induced by a Parasitic Nematode and a Rhizobial Endosymbiont. Mol. Plant-Microbe Interact. 2001, 14, 1168–1177. [Google Scholar] [CrossRef]
- Weerasinghe, R.R.; Bird, D.M.; Allen, N.S. Root-Knot Nematodes and Bacterial Nod Factors Elicit Common Signal Transduction Events in Lotus Japonicus. Proc. Natl. Acad. Sci. USA 2005, 102, 3147–3152. [Google Scholar] [CrossRef]
- Damiani, I.; Baldacci-Cresp, F.; Hopkins, J.; Andrio, E.; Balzergue, S.; Lecomte, P.; Puppo, A.; Abad, P.; Favery, B.; Hérouart, D. Plant Genes Involved in Harbouring Symbiotic Rhizobia or Pathogenic Nematodes. New Phytol. 2012, 194, 511–522. [Google Scholar] [CrossRef]
- Back, M.A.; Haydock, P.P.J.; Jenkinson, P. Disease Complexes Involving Plant Parasitic Nematodes and Soilborne Pathogens. Plant Pathol. 2002, 51, 683–697. [Google Scholar] [CrossRef]
- Molloy, B.; Baum, T.; Eves-van den Akker, S. Unlocking the Development- and Physiology-Altering ‘Effector Toolbox’ of Plant-Parasitic Nematodes. Trends Parasitol. 2023, 39, 732–738. [Google Scholar] [CrossRef]
- Pellegrin, C.; Damm, A.; Sperling, A.L.; Molloy, B.; Shin, D.S.; Long, J.; Brett, P.; Iguh, T.C.; Kranse, O.P.; Bravo, A.D.-T.; et al. The SUbventral-Gland Regulator (SUGR-1) of Nematode Virulence. Proc. Natl. Acad. Sci. USA 2025, 122, e2415861122. [Google Scholar] [CrossRef] [PubMed]
- Mabrouk, Y.; Hemissi, I.; Ben Salem, I.; Mejri, S.; Saidi, M.; Belhadj, O. Potential of Rhizobia in Improving Nitrogen Fixation and Yields of Legumes. In Symbiosis; Rigobelo, E., Ed.; IntechOpen: London, UK, 2018. [Google Scholar]
- Kanu, S.A.; Dakora, F.D. Symbiotic Functioning, Structural Adaptation, and Subcellular Organization of Root Nodules from Psoralea pinnata (L.) Plants Grown Naturally under Wetland and Upland Conditions in the Cape Fynbos of South Africa. Protoplasma 2017, 254, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.M.; Tegeder, M. Increasing Nitrogen Fixation and Seed Development in Soybean Requires Complex Adjustments of Nodule Nitrogen Metabolism and Partitioning Processes. Curr. Biol. 2016, 26, 2044–2051. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.; Tegeder, M. Soybean Ureide Transporters Play a Critical Role in Nodule Development, Function and Nitrogen Export. Plant J. 2012, 72, 355–367. [Google Scholar] [CrossRef]
- Redillas, M.C.F.R.; Bang, S.W.; Lee, D.; Kim, Y.S.; Jung, H.; Chung, P.J.; Suh, J.; Kim, J. Allantoin Accumulation through Overexpression of Ureide Permease1 Improves Rice Growth under Limited Nitrogen Conditions. Plant Biotechnol. J. 2019, 17, 1289–1301. [Google Scholar] [CrossRef]
- Gitari, H.I.; Nyawade, S.O.; Kamau, S.; Karanja, N.N.; Gachene, C.K.K.; Raza, M.A.; Maitra, S.; Schulte-Geldermann, E. Revisiting Intercropping Indices with Respect to Potato-Legume Intercropping Systems. Field Crops Res. 2020, 258, 107957. [Google Scholar] [CrossRef]
- Kebede, E. Contribution, Utilization, and Improvement of Legumes-Driven Biological Nitrogen Fixation in Agricultural Systems. Front. Sustain. Food Syst. 2021, 5, 767998. [Google Scholar] [CrossRef]
- Congreves, K.A.; Otchere, O.; Ferland, D.; Farzadfar, S.; Williams, S.; Arcand, M.M. Nitrogen Use Efficiency Definitions of Today and Tomorrow. Front. Plant Sci. 2021, 12, 637108. [Google Scholar] [CrossRef]
- Govindasamy, P.; Muthusamy, S.K.; Bagavathiannan, M.; Mowrer, J.; Jagannadham, P.T.K.; Maity, A.; Halli, H.M.; Sujayananad, G.K.; Vadivel, R.; Das, T.K.; et al. Nitrogen Use Efficiency—A Key to Enhance Crop Productivity under a Changing Climate. Front. Plant Sci. 2023, 14, 1121073. [Google Scholar] [CrossRef]
- Tamagno, S.; Maaz, T.M.; van Kessel, C.; Linquist, B.A.; Ladha, J.K.; Lundy, M.E.; Maureira, F.; Pittelkow, C.M. Critical Assessment of Nitrogen Use Efficiency Indicators: Bridging New and Old Paradigms to Improve Sustainable Nitrogen Management. Eur. J. Agron. 2024, 159, 127231. [Google Scholar] [CrossRef]
- Coggins, S.; McDonald, A.J.; Silva, J.V.; Urfels, A.; Nayak, H.S.; Sherpa, S.R.; Jat, M.L.; Jat, H.S.; Krupnik, T.; Kumar, V.; et al. Data-Driven Strategies to Improve Nitrogen Use Efficiency of Rice Farming in South Asia. Nat. Sustain. 2025, 8, 22–33. [Google Scholar] [CrossRef]
- Shadchina, T.M.; Dmitrieva, V.V. Leaf Chlorophyll Content as a Possible Diagnostic Mean for the Evaluation of Plant Nitrogen Uptake from the Soil. J. Plant Nutr. 1995, 18, 1427–1437. [Google Scholar] [CrossRef]
- Wang, D.; Xu, Z.; Zhao, J.; Wang, Y.; Yu, Z. Excessive Nitrogen Application Decreases Grain Yield and Increases Nitrogen Loss in a Wheat–Soil System. Acta Agric. Scand. B Soil. Plant Sci. 2011, 61, 681–692. [Google Scholar] [CrossRef]
- Brueck, H. Effects of Nitrogen Supply on Water-use Efficiency of Higher Plants. J. Plant Nutr. Soil. Sci. 2008, 171, 210–219. [Google Scholar] [CrossRef]
- Britto, D.T.; Kronzucker, H.J. NH4+ Toxicity in Higher Plants: A Critical Review. J. Plant Physiol. 2002, 159, 567–584. [Google Scholar] [CrossRef]
- Campos, C.N.S.; de Mello Prado, R.; Caione, G.; Neto, A.J.L.; Mingotte, F.L.C. Silicon and Excess Ammonium and Nitrate in Cucumber Plants. Afr. J. Agric. Res. 2016, 11, 276–283. [Google Scholar] [CrossRef][Green Version]
- Xia, X.; Ma, C.; Dong, S.; Xu, Y.; Gong, Z. Effects of Nitrogen Concentrations on Nodulation and Nitrogenase Activity in Dual Root Systems of Soybean Plants. Soil. Sci. Plant Nutr. 2017, 63, 470–482. [Google Scholar] [CrossRef]
- Abdel Wahab, A.M.; Zahran, H.H.; Abd-Alla, M.H. Root-Hair Infection and Nodulation of Four Grain Legumes as Affected by the Form and the Application Time of Nitrogen Fertilizer. Folia Microbiol. 1996, 41, 303–308. [Google Scholar] [CrossRef]
- Saito, A.; Tanabata, S.; Tanabata, T.; Tajima, S.; Ueno, M.; Ishikawa, S.; Ohtake, N.; Sueyoshi, K.; Ohyama, T. Effect of Nitrate on Nodule and Root Growth of Soybean (Glycine max (L.) Merr.). Int. J. Mol. Sci. 2014, 15, 4464–4480. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, X.; Xiao, J.; Tang, L.; Zheng, Y. Interactive Influences of Intercropping by Nitrogen on Flavonoid Exudation and Nodulation in Faba Bean. Sci. Rep. 2019, 9, 4818. [Google Scholar] [CrossRef]
- Ndakidemi, P.A.; Dakora, F.D. Legume Seed Flavonoids and Nitrogenous Metabolites as Signals and Protectants in Early Seedling Development. Funct. Plant Biol. 2003, 30, 729. [Google Scholar] [CrossRef]
- Akhtar, M.S.; Siddiqui, Z.A.; Wiemken, A. Arbuscular Mycorrhizal Fungi and Rhizobium to Control Plant Fungal Diseases. In Alternative Farming Systems, Biotechnology, Drought Stress and Ecological Fertilisation; Lichtfouse, E., Ed.; Springer: Dordrecht, The Netherlands, 2011; pp. 263–292. [Google Scholar]
- Godschalx, A.L.; Diethelm, A.C.; Kautz, S.; Ballhorn, D.J. Nitrogen-Fixing Rhizobia Affect Multitrophic Interactions in the Field. J. Insect Behav. 2023, 36, 168–179. [Google Scholar] [CrossRef]
- Gao, X.; Lu, X.; Wu, M.; Zhang, H.; Pan, R.; Tian, J.; Li, S.; Liao, H. Co-Inoculation with Rhizobia and AMF Inhibited Soybean Red Crown Rot: From Field Study to Plant Defense-Related Gene Expression Analysis. PLoS ONE 2012, 7, e33977. [Google Scholar] [CrossRef]
- Das, K.; Prasanna, R.; Saxena, A.K. Rhizobia: A Potential Biocontrol Agent for Soilborne Fungal Pathogens. Folia Microbiol. 2017, 62, 425–435. [Google Scholar] [CrossRef]
- Clúa, J.; Roda, C.; Zanetti, M.; Blanco, F. Compatibility between Legumes and Rhizobia for the Establishment of a Successful Nitrogen-Fixing Symbiosis. Genes 2018, 9, 125. [Google Scholar] [CrossRef]
- Soto, M.J.; Sanjuán, J.; Olivares, J. Rhizobia and Plant-Pathogenic Bacteria: Common Infection Weapons. Microbiology 2006, 152, 3167–3174. [Google Scholar] [CrossRef]
- Mabrouk, Y.; Belhadj, O. The Potential Use of Rhizobium–Legume Symbiosis for Enhancing Plant Growth and Management of Plant Diseases. In Microbes for Legume Improvement; Khan, M.S., Musarrat, J., Zaidi, A., Eds.; Springer: Vienna, Austria, 2010; pp. 495–514. [Google Scholar]
- Volpiano, C.G.; Lisboa, B.B.; Granada, C.E.; José, J.F.B.S.; de Oliveira, A.M.R.; Beneduzi, A.; Perevalova, Y.; Passaglia, L.M.P.; Vargas, L.K. Rhizobia for Biological Control of Plant Diseases. In Microbiome in Plant Health and Disease; Springer: Singapore, 2019; pp. 315–336. [Google Scholar]
- Parveen, G.; Noreen, R.; Shafique, H.A.; Sultana, V.; Ehteshamul-Haque, S.; Athar, M. Role of Rhizobia in Suppressing the Root Diseases of Soybean Under Soil Amendment. Planta Daninha 2019, 37, e019172336. [Google Scholar] [CrossRef]
- Khan, M.R.; Mohiddin, F.A.; Ahamad, F. Inoculant Rhizobia Suppressed Root-Knot Disease, and Enhanced Plant Productivity and Nutrient Uptake of Some Field-Grown Food Legumes. Acta Agric. Scand. B Soil. Plant Sci. 2018, 68, 166–174. [Google Scholar] [CrossRef]
- Omar, S.A.; Abd-Alla, M.H. Biocontrol of Fungal Root Rot Diseases of Crop Plants by the Use of Rhizobia and Bradyrhizobia. Folia Microbiol. 1998, 43, 431–437. [Google Scholar] [CrossRef]
- Akhtar, M.S.; Siddiqui, Z.A. Biocontrol of a Root-Rot Disease Complex of Chickpea by Glomus Intraradices, Rhizobium Sp. and Pseudomonas Straita. Crop Prot. 2008, 27, 410–417. [Google Scholar] [CrossRef]
- Castillo, P.; Navas-Cortés, J.A.; Landa, B.B.; Jiménez-Díaz, R.M.; Vovlas, N. Plant-Parasitic Nematodes Attacking Chickpea and Their In Planta Interactions with Rhizobia and Phytopathogenic Fungi. Plant Dis. 2008, 92, 840–853. [Google Scholar] [CrossRef]
- Noreen, R.; Shafique, A.; Ali, S.A.; Habiba, H.; Sultana, V.; Ara, J.; Ehteshamul-Haque, S. Role of Mungbean Root Nodule Associated Fluorescent Pseudomonas and Rhizobia in Suppressing the Root Rotting Fungi and Root Knot Nematodes in Chickpea (Cicer arietinum L.). Pak. J. Bot. 2016, 48, 2139–2145. [Google Scholar]
- Tamiru, G.; Muleta, D. The Effect of Rhizobia Isolates Against Black Root Rot Disease of Faba Bean (Vicia faba L) Caused by Fusarium Solani. Open Agric. J. 2018, 12, 131–147. [Google Scholar] [CrossRef]
- Dean, J.; Mescher, M.; De Moraes, C. Plant Dependence on Rhizobia for Nitrogen Influences Induced Plant Defenses and Herbivore Performance. Int. J. Mol. Sci. 2014, 15, 1466–1480. [Google Scholar] [CrossRef]
- Thamer, S.; Schädler, M.; Bonte, D.; Ballhorn, D.J. Dual Benefit from a Belowground Symbiosis: Nitrogen Fixing Rhizobia Promote Growth and Defense against a Specialist Herbivore in a Cyanogenic Plant. Plant Soil. 2011, 341, 209–219. [Google Scholar] [CrossRef]
- Deshwal, V.K.; Pandey, P.; Kang, S.C.; Maheshwari, D.K. Rhizobia as a Biological Control Agent against Soil Borne Plant Pathogenic Fungi. Indian J. Exp. Biol. 2003, 41, 1160–1164. [Google Scholar]
- Hossain, M.M.; Sultana, F.; Yesmin, L.; Rubayet, M.T.; Abdullah, H.M.; Siddique, S.S.; Bhuiyan, M.A.B.; Yamanaka, N. Understanding Phakopsora Pachyrhizi in Soybean: Comprehensive Insights, Threats, and Interventions from the Asian Perspective. Front. Microbiol. 2024, 14, 1304205. [Google Scholar] [CrossRef]
- Van Haeften, S.; Kang, Y.; Dudley, C.; Potgieter, A.; Robinson, H.; Dinglasan, E.; Wenham, K.; Noble, T.; Kelly, L.; Douglas, C.A.; et al. Fusarium Wilt Constrains Mungbean Yield Due to Reduction in Source Availability. AoB Plants 2024, 16, plae021. [Google Scholar] [CrossRef]
- Arya, A.; Mishra, P.; Yadav, A.; Singh, A.; Kumar, A. Collar Rot Disease of Lentil Caused by Sclerotium Rolfsii and Its Management. J. Pharmacogn. Phytochem. 2021, 10, 1012–1016. [Google Scholar]
- Praveen, A.; Kannan, C. Disease Incidence and Severity of Sclerotium Rolfsii on Arachis hypogea L. Plant Arch. 2021, 21, 344–349. [Google Scholar] [CrossRef]
- Mengesha, G.G.; Terefe, H.; Cheleko, D.C. Progression of Chocolate Spot (Botrytis Fabae) and Grain Yield of Faba Bean as Influenced by Integration of Fungicide Rate and Host Resistance in Southern Ethiopia. J. Crop Sci. Biotechnol. 2022, 25, 73–90. [Google Scholar] [CrossRef]
- Iqbal, U.; Iqbal, S.M.; Afzal, R.; Jamal, A.; Farooq, M.A.; Zahid, A. Screening of Mungbean Germplasm against Mungbean Yellow Mosaic Virus (MYMV) under Field Conditions. Pak. J. Phytopathol. 2011, 23, 48–51. [Google Scholar]
- Jha, U.C.; Nayyar, H.; Chattopadhyay, A.; Beena, R.; Lone, A.A.; Naik, Y.D.; Thudi, M.; Prasad, P.V.V.; Gupta, S.; Dixit, G.P.; et al. Major Viral Diseases in Grain Legumes: Designing Disease Resistant Legumes from Plant Breeding and OMICS Integration. Front. Plant Sci. 2023, 14, 1183505. [Google Scholar] [CrossRef]
- Mushadu, P.N. Viral Diseases of Legumes and Their Managements. In Advances in Legume Research: Physiological Responses and Genetic Improvement for Biotic Stress Resistance; Bentham Science Publishers: Sharjah, United Arab Emirates, 2023; Volume 2, pp. 64–82. [Google Scholar]
- Fininsa, C.; Yuen, J. Association of Bean Rust and Common Bacterial Blight Epidemics with Cropping Systems in Hararghe Highlands, Eastern Ethiopia. Int. J. Pest. Manag. 2001, 47, 211–219. [Google Scholar] [CrossRef]
- Karavina, C.; Mandumbu, R.; Parwada, C.; Zivenge, E. Epiphytic Survival of Xanthomonas Axonopodis Pv. Phaseoli (EF SM). J. Anim. Plant Sci. 2011, 9, 1161–1168. [Google Scholar]
- Belete, T.; Bastas, K.K. Common Bacterial Blight (Xanthomonas axonopodis pv. phaseoli) of Beans with Special Focus on Ethiopian Condition. J. Plant Pathol. Microbiol. 2017, 8, 1000403. [Google Scholar] [CrossRef]
- Gao, L.; Sun, S.; Li, K.; Wang, L.; Hou, W.; Wu, C.; Zhi, H.; Han, T. Spatio-Temporal Characterisation of Changes in the Resistance of Widely Grown Soybean Cultivars to Soybean Mosaic Virus across a Century of Breeding in China. Crop Pasture Sci. 2018, 69, 395. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Coutts, B.A.; Latham, L.J.; McKirdy, S.J. Cucumber Mosaic Virus Infection of Chickpea Stands: Temporal and Spatial Patterns of Spread and Yield-limiting Potential. Plant Pathol. 2008, 57, 842–853. [Google Scholar] [CrossRef]
- Foresto, E.; Carezzano, M.E.; Giordano, W.; Bogino, P. Ascochyta Blight in Chickpea: An Update. J. Fungi 2023, 9, 203. [Google Scholar] [CrossRef]
- Iqbal, J.; Yousaf, U.; Zia, S.; Asgher, A.; Afzal, R.; Ali, M.; Sheikh, A.U.R.; Sher, A. Pulses Diseases “Important Limiting Factor in Yield’’ and Their Managements. Asian J. Res. Crop Sci. 2019, 3, 1–21. [Google Scholar] [CrossRef]
- Ijaz, U.; Adhikari, K.; Kimber, R.; Trethowan, R.; Bariana, H.; Bansal, U. Pathogenic Specialization in Uromyces Viciae-Fabae in Australia and Rust Resistance in Faba Bean. Plant Dis. 2021, 105, 636–642. [Google Scholar] [CrossRef]
- Upadhyay, V.; Medhi, K.; Pandey, P.; Thengal, P.; Paul, S.; Kushwaha, K.P. Rust Disease of Pea: A Review. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 416–434. [Google Scholar] [CrossRef]
- Kuchuran, M.E.; Armstrong-Cho, C.; Banniza, S. Management of Botrytis Grey Mould Caused by Botrytis Cinerea in Lentil Using Boscalid Fungicide. Can. J. Plant Pathol. 2021, 43, 694–703. [Google Scholar] [CrossRef]
- Adetunji, M.C.; Alika, O.P.; Awa, N.P.; Atanda, O.O.; Mwanza, M. Microbiological Quality and Risk Assessment for Aflatoxins in Groundnuts and Roasted Cashew Nuts Meant for Human Consumption. J. Toxicol. 2018, 2018, 1308748. [Google Scholar] [CrossRef]
- Tumsa, K.; Shimelis, H.; Laing, M.; Mukankusi, C.; Mathew, I. Identification of Sources of Resistance to Common Bacterial Blight in Common Bean in Ethiopia. J. Phytopathol. 2020, 168, 707–720. [Google Scholar] [CrossRef]
- Adila, W.; Terefe, H.; Bekele, A. Common Bacterial Blight (Xanthomonas axonopodis pv. phaseoli) Resistance Reaction in Common Bean Genotypes and Their Agronomic Performances in Southern Ethiopia. J. Crop Sci. Biotechnol. 2021, 24, 387–400. [Google Scholar] [CrossRef]
- Foyer, C.H.; Lam, H.-M.; Nguyen, H.T.; Siddique, K.H.M.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M.; et al. Neglecting Legumes Has Compromised Human Health and Sustainable Food Production. Nat. Plants 2016, 2, 16112. [Google Scholar] [CrossRef]
- Maitlo, S.A.; Syed, R.N.; Rustamani, M.A.; Khuhro, R.D.; Lodhi, A.M. Comparative Efficacy of Different Fungicides against Fusarium Wilt of Chickpea (Cicer arietinum L.). Pak. J. Bot. 2014, 46, 2305–2312. [Google Scholar]
- Sbai, H.; Hajib, A.; Msairi, S.; Amalich, S.; Bouyahya, A.; Lee, L.-H.; Goh, K.W.; Tabyaoui, M.; Harhar, H. Fungal Infections of Legume Crops: Challenges and Management Approaches. J. Agric. Food Res. 2024, 18, 101447. [Google Scholar] [CrossRef]
- Bueno, M.R.; Cunha, J.P.A.D. Environmental Risk for Aquatic and Terrestrial Organisms Associated with Drift from Pesticides Used in Soybean Crops. An. Acad. Bras. Cienc. 2020, 92, e20181245. [Google Scholar] [CrossRef]
- Hilber, I.; Bahena-Juárez, F.; Chiaia-Hernández, A.C.; Elgueta, S.; Escobar-Medina, A.; Friedrich, K.; González-Curbelo, M.Á.; Grob, Y.; Martín-Fleitas, M.; Miglioranza, K.S.B.; et al. Pesticides in Soil, Groundwater and Food in Latin America as Part of One Health. Environ. Sci. Pollut. Res. 2024, 31, 14333–14345. [Google Scholar] [CrossRef]
- Zubrod, J.P.; Bundschuh, M.; Arts, G.; Brühl, C.A.; Imfeld, G.; Knäbel, A.; Payraudeau, S.; Rasmussen, J.J.; Rohr, J.; Scharmüller, A.; et al. Fungicides: An Overlooked Pesticide Class? Environ. Sci. Technol. 2019, 53, 3347–3365. [Google Scholar] [CrossRef] [PubMed]
- Derbalah, A.; Shebl, A.M.; Elgobashy, S.F.; Ahmad, A.A.; Ramadan, N.E.; Behiry, S.I.; Abdelkhalek, A.; Saleem, M.H.; Al-Askar, A.A.; Kamran, M.; et al. Resistance Induction and Direct Antifungal Activity of Some Monoterpenes against Rhizoctonia Solani, the Causal of Root Rot in Common Bean. Life 2022, 12, 1040. [Google Scholar] [CrossRef] [PubMed]
- Ceresini, P.C.; Silva, T.C.; Vicentini, S.N.C.; Júnior, R.P.L.; Moreira, S.I.; Castro-Ríos, K.; Garcés-Fiallos, F.R.; Krug, L.D.; de Moura, S.S.; da Silva, A.G.; et al. Strategies for Managing Fungicide Resistance in the Brazilian Tropical Agroecosystem: Safeguarding Food Safety, Health, and the Environmental Quality. Trop. Plant Pathol. 2024, 49, 36–70. [Google Scholar] [CrossRef]
- Gorshkov, A.P.; Kusakin, P.G.; Borisov, Y.G.; Tsyganova, A.V.; Tsyganov, V.E. Effect of Triazole Fungicides Titul Duo and Vintage on the Development of Pea (Pisum sativum L.) Symbiotic Nodules. Int. J. Mol. Sci. 2023, 24, 8646. [Google Scholar] [CrossRef]
- A/HRC/34/48; United Nations Report of the Secretary-General on the Right to Food. WFP: Rome, Italy, 2017.
- A/RES/68/231; The International Year of Pulses Final Report. FAO: Rome, Italy, 2016. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/4ac63815-9b93-4103-b3e2-71521aa30857/content (accessed on 3 July 2025).
- Semba, R.D.; Ramsing, R.; Rahman, N.; Kraemer, K.; Bloem, M.W. Legumes as a Sustainable Source of Protein in Human Diets. Glob. Food Secur. 2021, 28, 100520. [Google Scholar] [CrossRef]
- Vilakazi, B.; Mafongoya, P.L.; Odindo, A.O.; Phophi, M.M. The Role of Neglected Grain Legumes in Food and Nutrition Security and Human Health. Sustainability 2025, 17, 350. [Google Scholar] [CrossRef]
- Arce, C.E.; Caballero, J. Tanzania: Agricultural Sector Risk Assessment; World Bank: Washington, DC, USA, 2015; pp. 1–79. [Google Scholar]
- Breen, C.; Ndlovu, N.; McKeown, P.C.; Spillane, C. Legume Seed System Performance in Sub-Saharan Africa: Barriers, Opportunities, and Scaling Options. A Review. Agron. Sustain. Dev. 2024, 44, 20. [Google Scholar] [CrossRef]
- Riccioni, L.; Orzali, L.; Romani, M.; Annicchiarico, P.; Pecetti, L. Organic Seed Treatments with Essential Oils to Control Ascochyta Blight in Pea. Eur. J. Plant Pathol. 2019, 155, 831–840. [Google Scholar] [CrossRef]
- Schreinemachers, P.; Balasubramaniam, S.; Boopathi, N.M.; Ha, C.V.; Kenyon, L.; Praneetvatakul, S.; Sirijinda, A.; Le, N.T.; Srinivasan, R.; Wu, M.-H. Farmers’ Perceptions and Management of Plant Viruses in Vegetables and Legumes in Tropical and Subtropical Asia. Crop Prot. 2015, 75, 115–123. [Google Scholar] [CrossRef]
- Ndabashinze, B.; Nchanji, E.B.; Lutomia, C.K.; Nduwarugira, E.; Hakizimana, M.B.; Mayugi, I. Closing Gender Gaps through Gender-Responsive, Demand-Led Breeding in Burundi. Front. Sociol. 2024, 8, 1264816. [Google Scholar] [CrossRef]
- Nchanji, E.B.; Lutomia, C.K.; Ageyo, O.C.; Karanja, D.; Kamau, E. Gender-Responsive Participatory Variety Selection in Kenya: Implications for Common Bean (Phaseolus vulgaris L.) Breeding in Kenya. Sustainability 2021, 13, 13164. [Google Scholar] [CrossRef]
- Nchanji, E. Women Making Great Strides in Ensuring Food Security in Burundi. Available online: https://alliancebioversityciat.org/stories/women-making-great-strides-ensuring-food-security-burundi (accessed on 3 July 2025).
- Nchanji, E.; Acheampong, P.; Ngoh, S.B.; Nyamolo, V.; Cosmas, L. Comparative Analysis of Youth Transition in Bean Production Systems in Ghana and Cameroon. Humanit. Soc. Sci. Commun. 2024, 11, 154. [Google Scholar] [CrossRef]
- Dessalegn, B.; Asnake, W.; Tigabie, A.; Le, Q.B. Challenges to Adoption of Improved Legume Varieties: A Gendered Perspective. Sustainability 2022, 14, 2150. [Google Scholar] [CrossRef]
- Sheeba, J.R.; Raja, B.C. Integrated Pest and Disease Management Module in Groundnut (Arachis hypogaea L.) at Tiruchirappalli District, Tamil Nadu, India. J. Adv. Biol. Biotechnol. 2025, 28, 656–660. [Google Scholar] [CrossRef]
- Karthikeyan, M.; Dhanabalan, S.P.; Shanmugavel, B.; Jaffer, S.B.; Marimuthu, S.; Radhika, K.; Nagappan, E.; Johnson, I.; Periyannan, S. Aflatoxin Evaluation and Integrated Management Strategies to Minimize Toxin Contamination in Maize and Peanuts. J. Agric. Food Res. 2025, 21, 101809. [Google Scholar] [CrossRef]
- Gremillion, S.; Culbreath, A.; Gorbet, D.; Mullinix, B.; Pittman, R.; Stevenson, K.; Todd, J.; Condori, M. Response of Progeny Bred from Bolivian and North American Cultivars in Integrated Management Systems for Leaf Spot of Peanut (Arachis hypogaea). Crop Prot. 2011, 30, 698–704. [Google Scholar] [CrossRef]
- Cantonwine, E.G.; Culbreath, A.K.; Stevenson, K.L.; Kemerait, R.C.; Brenneman, T.B.; Smith, N.B.; Mullinix, B.G. Integrated Disease Management of Leaf Spot and Spotted Wilt of Peanut. Plant Dis. 2006, 90, 493–500. [Google Scholar] [CrossRef]
- Kodape, A.; Lama, A.; Vivek Babu, C.S. Metagenomic Insights of Fungal Diversity of Peanuts under Storage Conditions and Mitigation of Aflatoxigenic Fungi through Competitive Exclusion and Phytochemicals. Food Biosci. 2024, 58, 103711. [Google Scholar] [CrossRef]
- Rahman, M.T.; Rubayet, M.T.; Khan, A.A.; Bhuiyan, M.K.A. Integrated Management of Fusarium Root Rot and Wilt Disease of Soybean Caused by Fusarium oxysporum. Int. J. Biosci. 2020, 17, 83–96. [Google Scholar] [CrossRef]
- Siddiqui, Z.S.; Nida, K.; Cho, J.-I.; Rehman, Y.; Abideen, Z. Physiological and Photochemical Profiling of Soybean Plant Using Biological and Chemical Methods of Treatment against Biotic Stress Management. Plant Physiol. Biochem. 2024, 208, 108454. [Google Scholar] [CrossRef]
- Estevez de Jensen, C.; Kurle, J.E.; Percich, J.A. Integrated Management of Edaphic and Biotic Factors Limiting Yield of Irrigated Soybean and Dry Bean in Minnesota. Field Crops Res. 2004, 86, 211–224. [Google Scholar] [CrossRef]
- Rahman, M.T.; Rubayet, M.T.; Khan, A.A.; Bhuiyan, M.K.A. Integrated Management of Charcoal Rot Disease of Soybean Caused by Macrophomina Phaseolina. Egypt. J. Agric. Res. 2021, 99, 10–19. [Google Scholar] [CrossRef]
- Pedersen, P.; Grau, C.; Cullen, E.; Koval, N.; Hill, J.H. Potential for Integrated Management of Soybean Virus Disease. Plant Dis. 2007, 91, 1255–1259. [Google Scholar] [CrossRef] [PubMed]
- Culbreath, A.K.; Selph, A.C.; Williams, B.W.; Kemerait, R.C.; Srinivasan, R.; Abney, M.R.; Tillman, B.L.; Holbrook, C.C.; Branch, W.D. Effects of New Field Resistant Cultivars and In-Furrow Applications of Phorate Insecticide on Tomato Spotted Wilt of Peanut. Crop Prot. 2016, 81, 70–75. [Google Scholar] [CrossRef]
- Kaur, L.; Campbell, H.L.; Miller, H.B.; Parker, C.; Burkett, J.; Strayer-Scherer, A.L. Response of Selected Peanut Commercial Cultivars to Leaf Spot Diseases as Influenced by Fungicide Inputs. Crop Prot. 2024, 184, 106781. [Google Scholar] [CrossRef]
- Culbreath, A.K.; Tubbs, R.S.; Tillman, B.L.; Beasley, J.P.; Branch, W.D.; Holbrook, C.C.; Smith, A.R.; Smith, N.B. Effects of Seeding Rate and Cultivar on Tomato Spotted Wilt of Peanut. Crop Prot. 2013, 53, 118–124. [Google Scholar] [CrossRef]
- Dorrance, A.E.; Robertson, A.E.; Cianzo, S.; Giesler, L.J.; Grau, C.R.; Draper, M.A.; Tenuta, A.U.; Anderson, T.R. Integrated Management Strategies for Phytophthora Sojae Combining Host Resistance and Seed Treatments. Plant Dis. 2009, 93, 875–882. [Google Scholar] [CrossRef]
- Brown, M.T.; Mueller, D.S.; Kandel, Y.R.; Telenko, D.E.P. Influence of Integrated Management Strategies on Soybean Sudden Death Syndrome (SDS) Root Infection, Foliar Symptoms, Yield and Net Returns. Pathogens 2023, 12, 913. [Google Scholar] [CrossRef]
- Hu, H.; Zhao, J.; Thomas, W.J.W.; Batley, J.; Edwards, D. The Role of Pangenomics in Orphan Crop Improvement. Nat. Commun. 2025, 16, 118. [Google Scholar] [CrossRef]
- Bhadauria, V.; Ramsay, L.; Bett, K.E.; Banniza, S. QTL Mapping Reveals Genetic Determinants of Fungal Disease Resistance in the Wild Lentil Species Lens Ervoides. Sci. Rep. 2017, 7, 3231. [Google Scholar] [CrossRef]
- Kankanala, P.; Nandety, R.S.; Mysore, K.S. Genomics of Plant Disease Resistance in Legumes. Front. Plant Sci. 2019, 10, 1345. [Google Scholar] [CrossRef]
- Anglin, N.L.; Amri, A.; Kehel, Z.; Ellis, D. A Case of Need: Linking Traits to Genebank Accessions. Biopreserv. Biobank. 2018, 16, 337–349. [Google Scholar] [CrossRef]
- Isleib, T.G.; Holbrook, C.C.; Gorbet, D.W. Use of Plant Introductions in Peanut Cultivar Development. Peanut Sci. 2001, 28, 96–113. [Google Scholar] [CrossRef]
- Sharma, S.; Upadhyaya, H.D.; Varshney, R.K.; Gowda, C.L.L. Pre-Breeding for Diversification of Primary Gene Pool and Genetic Enhancement of Grain Legumes. Front. Plant Sci. 2013, 4, 309. [Google Scholar] [CrossRef]
- Mahuku, G.S.; Jara, C.; Cajiao, C.; Beebe, S. Sources of Resistance to Angular Leaf Spot (Phaeoisariopsis griseola) in Common Bean Core Collection, Wild Phaseolus vulgaris and Secondary Gene Pool. Euphytica 2003, 130, 303–313. [Google Scholar] [CrossRef]
- Gayacharan; Parida, S.K.; Mondal, N.; Yadav, R.; Vishwakarma, H.; Rana, J.C. Mining Legume Germplasm for Genetic Gains: An Indian Perspective. Front. Genet. 2023, 14, 996828. [Google Scholar] [CrossRef]
- Varshney, R.K.; Roorkiwal, M.; Sun, S.; Bajaj, P.; Chitikineni, A.; Thudi, M.; Singh, N.P.; Du, X.; Upadhyaya, H.D.; Khan, A.W.; et al. A Chickpea Genetic Variation Map Based on the Sequencing of 3366 Genomes. Nature 2021, 599, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Contreras, M.P.; Pai, H.; Selvaraj, M.; Toghani, A.; Lawson, D.M.; Tumtas, Y.; Duggan, C.; Yuen, E.L.H.; Stevenson, C.E.M.; Harant, A.; et al. Resurrection of Plant Disease Resistance Proteins via Helper NLR Bioengineering. Sci. Adv. 2023, 9, eadg3861. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Zhang, Q.; Lin, Y.; Chen, J.; Somta, P.; Yan, Q.; Xue, C.; Liu, J.; Chen, X.; Yuan, X. Marker-Assisted Backcross Breeding for Improving Bruchid (Callosobruchus spp.) Resistance in Mung Bean (Vigna radiata L.). Agronomy 2022, 12, 1271. [Google Scholar] [CrossRef]
- Kiryowa, M.J.; Nkalubo, S.T.; Mukankusi, C.; Male, A.; Tukamuhabwe, P.; Rubaihayo, P.; Gibson, P.T. Effectiveness of Pyramided Genes in Conferring Resistance to Anthracnose Disease in Common Bean Populations. J. Plant Breed. Crop Sci. 2021, 13, 1–13. [Google Scholar] [CrossRef]
- Lv, Z.; Lan, G.; Bai, B.; Yu, P.; Wang, C.; Zhang, H.; Zhong, C.; Zhao, X.; Yu, H. Identification of Candidate Genes Associated with Peanut Pod Length by Combined Analysis of QTL-Seq and RNA-Seq. Genomics 2024, 116, 110835. [Google Scholar] [CrossRef]
- Wang, F.; Miao, H.; Zhang, S.; Hu, X.; Li, C.; Yang, W.; Chen, J. Identification of a New Major Oil Content QTL Overlapped with FAD2B in Cultivated Peanut (Arachis hypogaea L.). Plants 2025, 14, 615. [Google Scholar] [CrossRef]
- Ravelombola, W.; Qin, J.; Shi, A.; Song, Q.; Yuan, J.; Wang, F.; Chen, P.; Yan, L.; Feng, Y.; Zhao, T.; et al. Genome-Wide Association Study and Genomic Selection for Yield and Related Traits in Soybean. PLoS ONE 2021, 16, e0255761. [Google Scholar] [CrossRef]
- Chen, Y.; Xiong, H.; Ravelombola, W.; Bhattarai, G.; Barickman, C.; Alatawi, I.; Phiri, T.M.; Chiwina, K.; Mou, B.; Tallury, S.; et al. A Genome-Wide Association Study Reveals Region Associated with Seed Protein Content in Cowpea. Plants 2023, 12, 2705. [Google Scholar] [CrossRef]
- Keller, B.; Ariza-Suarez, D.; de la Hoz, J.; Aparicio, J.S.; Portilla-Benavides, A.E.; Buendia, H.F.; Mayor, V.M.; Studer, B.; Raatz, B. Genomic Prediction of Agronomic Traits in Common Bean (Phaseolus vulgaris L.) Under Environmental Stress. Front. Plant Sci. 2020, 11, 1001. [Google Scholar] [CrossRef] [PubMed]
- Taku, M.; Saini, M.; Kumar, R.; Debbarma, P.; Rathod, N.K.K.; Onteddu, R.; Sharma, D.; Pandey, R.; Gaikwad, K.; Lal, S.K.; et al. Modified Speed Breeding Approach Reduced Breeding Cycle to Less than Half in Vegetable Soybean [Glycine max (L.) Merr.]. Physiol. Mol. Biol. Plants 2024, 30, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Habde, S.V.; Punniyamoorthy, D.; Jegadeesan, S. Mutation Profiling through Whole Genome Sequencing of Electron Beam-Induced Black Gram (Vigna mungo L. Hepper) Mutant. Int. J. Radiat. Biol. 2024, 100, 1665–1682. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Qu, S.; Liu, F.; Sun, H.; Li, H.; Teng, W.; Zhan, Y.; Li, Y.; Han, Y.; Zhao, X. Multi-omics Analysis Identified the GmUGT88A1 Gene, Which Coordinately Regulates Soybean Resistance to Cyst Nematode and Isoflavone Content. Plant Biotechnol. J. 2025, 23, 1291–1307. [Google Scholar] [CrossRef] [PubMed]
- Hodge, B.; Batnini, A.; Bolaños-Carriel, C.; Van, K.; Maroof, M.A.S.; McHale, L.; Dorrance, A.E. Resistance Gene Enrichment Sequencing for NLR Genes for Phytophthora Sojae in Selected Soybean Plant Introductions and Differentials with Putative Novel and Known Rps Genes. Crop Sci. 2025, 65, e21413. [Google Scholar] [CrossRef]
- Choudhury, A.; Rajam, M.V. Genetic Transformation of Legumes: An Update. Plant Cell Rep. 2021, 40, 1813–1830. [Google Scholar] [CrossRef]
- Vargas-Almendra, A.; Ruiz-Medrano, R.; Núñez-Muñoz, L.A.; Ramírez-Pool, J.A.; Calderón-Pérez, B.; Xoconostle-Cázares, B. Advances in Soybean Genetic Improvement. Plants 2024, 13, 3073. [Google Scholar] [CrossRef]
- ISAAA International Service for the Acquisition of Agri-Biotech Applications. Available online: https://www.isaaa.org/gmapprovaldatabase/eventslist/default.asp (accessed on 3 July 2025).
- Tripathi, A.; Rathore, M.; Shukla, S.; Das, A.; Debnath, S.C. Agrobacterium and Biolistic Mediated Genetic Transformation of Mungbean Cultivar Samrat Using Embryogenic Explant. Plant Cell Tissue Organ Cult. 2024, 157, 72. [Google Scholar] [CrossRef]
- Wang, T.; Xun, H.; Wang, W.; Ding, X.; Tian, H.; Hussain, S.; Dong, Q.; Li, Y.; Cheng, Y.; Wang, C.; et al. Mutation of GmAITR Genes by CRISPR/Cas9 Genome Editing Results in Enhanced Salinity Stress Tolerance in Soybean. Front. Plant Sci. 2021, 12, 779598. [Google Scholar] [CrossRef]
- Rajyaguru, R.H.; Tomar, R.S. Induction of New Allelic Variant of AhFAD2B Gene in Peanut Cultivar, GG20 through CRISPR/Cas9-Mediated Mutagenesis. J. Plant Biochem. Biotechnol. 2024, 33, 248–254. [Google Scholar] [CrossRef]
- Gao, L.; Xie, L.; Xiao, Y.; Cheng, X.; Pu, R.; Zhang, Z.; Liu, Y.; Gao, S.; Zhang, Z.; Qu, H.; et al. CRISPR/CasRx-Mediated Resistance to Soybean Mosaic Virus in Soybean. Crop J. 2024, 12, 1093–1101. [Google Scholar] [CrossRef]
- Dhobale, K.V.; Sahoo, L. Hairpin-RNA Spray Confers Resistance to Mungbean Yellow Mosaic India Virus in Mungbean. New Results 2024, 3, 585278. [Google Scholar]
- Mukankusi, C.; Raatz, B.; Nkalubo, S.; Berhanu, F.; Binagwa, P.; Kilango, M.; Williams, M.; Enid, K.; Chirwa, R.; Beebe, S. Genomics, Genetics and Breeding of Common Bean in Africa: A Review of Tropical Legume Project. Plant Breed. 2019, 138, 401–414. [Google Scholar] [CrossRef]
- Vanlauwe, B.; Hungria, M.; Kanampiu, F.; Giller, K.E. The Role of Legumes in the Sustainable Intensification of African Smallholder Agriculture: Lessons Learnt and Challenges for the Future. Agric. Ecosyst. Environ. 2019, 284, 106583. [Google Scholar] [CrossRef]
- Lisciani, S.; Marconi, S.; Le Donne, C.; Camilli, E.; Aguzzi, A.; Gabrielli, P.; Gambelli, L.; Kunert, K.; Marais, D.; Vorster, B.J.; et al. Legumes and Common Beans in Sustainable Diets: Nutritional Quality, Environmental Benefits, Spread and Use in Food Preparations. Front. Nutr. 2024, 11, 1385232. [Google Scholar] [CrossRef]
- Ojiewo, C.O.; Omoigui, L.O.; Pasupuleti, J.; Lenné, J.M. Grain Legume Seed Systems for Smallholder Farmers: Perspectives on Successful Innovations. Outlook Agric. 2020, 49, 286–292. [Google Scholar] [CrossRef]
- Tripodi, P.; Nicastro, N.; Pane, C. Digital Applications and Artificial Intelligence in Agriculture toward Next-Generation Plant Phenotyping. Crop Pasture Sci. 2022, 74, 597–614. [Google Scholar] [CrossRef]
| Crop | Disease | Causal Agent | Major Countries Affected | Reported Yield Loss (%) |
|---|---|---|---|---|
| Common Bean | Common Bacterial Blight | Xanthomonas spp. | USA, Brazil, Kenya | Up to 45% |
| Anthracnose | Colletotrichum lindemuthianum | India, USA, Brazil | 20–60% | |
| Halo Blight | Pseudomonas syringae pv. phaseolicola | Ethiopia, East Africa | Up to 45% | |
| Soybean | Bacterial Blight | Pseudomonas syringae pv. glycinea | USA, Argentina, China | 4–40% |
| Phytophthora Root Rot | Phytophthora spp. | USA, Brazil, China | Up to 50% | |
| Downy mildew | Peronospora spp. | USA, Central Europe, Asia, Africa | 5–10% | |
| Chickpea | Ascochyta Blight | Ascochyta rabiei | India, Australia, Turkey | Up to 100% |
| Fusarium Wilt | Fusarium oxysporum f. sp. ciceris | India, Pakistan, Spain | 10–90% | |
| Field Pea | Ascochyta Blight | Ascochyta pisi, A. pinodes, A. pinodella | Canada, Australia, UK | 20–50% |
| Downy Mildew | Peronospora viciae | USA, UK, India | 10–40% | |
| Bacterial Blight | Pseudomonas syringae pv. pisi | Ethiopia, Australia, UK, Canada, US | Up to 22% | |
| Root Rot | Aphanomyces euteiches | Ethiopia, US, Canada, France | 22–80% | |
| Cowpea | Cercospora Leaf Spot | Mycosphaerella cruenta | Nigeria, India, China | Up to 40% |
| Lentil | Root Rot Complex | Rhizoctonia solani, Fusarium spp., Pythium spp. | Canada, India, Nepal | 10–30% |
| Ascochyta blight | Ascochyta lentis | China, Italy | 23–62% | |
| Stemphylium blight | Stemphylium botryosum | Bangladesh, Canada, Ethiopia, Morocco, Syria | Up to 100% | |
| Fusarium wilt | Fusarium oxysporum f.sp. lentis | India, West Asia, North Africa, East Africa, Syria, Pakistan, Czechoslovakia | 40–90% | |
| Anthracnose | Colletotrichum truncatum | Bangladesh, Canada, Ethiopia, Morocco, Syria | 23–62% | |
| Rust | Uromyces fabae | Bangladesh, Canada, Ethiopia, Morocco, Syria | Up to 100% | |
| Faba bean | Ascochyta blight | Ascochyta fabae | Australia | 30–70% |
| Chocolate spot | Botrytis fabae | China, Ethiopia, Australia | Up to 100% | |
| Rust | Uromyces viciae-fabae | Bangladesh, Canada, Ethiopia, Morocco, Syria | 27–80% | |
| Black root rot | Fusarium solani | Bangladesh, Canada, Ethiopia, Morocco, Syria | Up to 100% |
| Genus | Virus | Major Tropical Legume Hosts | Symptoms | Transmitted Through | References |
|---|---|---|---|---|---|
| Alfamovirus | Alfalfa mosaic virus | Soybean | Leaf mottling; light and dark green, yellow patches; leaf curling; deformation; stunting | Aphids (plant lice); seeds or pollen to the seed | [62] |
| Begomovirus | Bean golden mosaic virus | Common bean, Lima bean | Yellow-green mosaic patterns on leaves; distorted and stunted plant growth | Whiteflies (Bemisia tabaci) | [63] |
| Bean golden yellow mosaic virus | Common bean | Yellow-green mosaic patterns on leaves; distorted and stunted plant growth | Whiteflies (Bemisia tabaci) | [64] | |
| Dolichos yellow mosaic virus | Lablab bean | Mosaic yellow pattern; patches of yellow alternating with green; stunting; leaf curling; reduced pod size | Whiteflies (Bemisia tabaci); seed transmission | [65] | |
| Horsegram yellow mosaic virus | Common bean, Mung bean, Pigeon pea | Bright yellow mosaic patterns on the leaves; reduced leaf size; rugosity; stunting of the entire plant | Whiteflies (Bemisia tabaci) | [66] | |
| Macroptilium yellow spot virus | Common bean, Lima bean | Yellowing or spotting on the leaves; bright yellow mosaic patterns on the leaves; a combination of bright yellow and green patches; stunted growth | Whiteflies (Bemisia tabaci) | [67] | |
| Mungbean yellow mosaic virus | Mung bean, Soybean, Common bean, Cowpea, Black gram, Pigeon pea | Yellow mosaic patterns, leaf curling, stunting | Whiteflies (Bemisia tabaci) | [68] | |
| Mungbean yellow mosaic India virus | Mung bean, Soybean, Common bean, Cowpea, Black gram, Lima bean, Pigeon pea, Lablab bean | Bright yellow mosaic patterns on the leaves; stunted growth; reduced leaf size; shriveled and misshapen seeds | Whiteflies (Bemisia tabaci) | [69] | |
| Tomato leaf curl virus | Soybean, Common bean | Stunting; reduced leaf size; upward curling of leaves; interveinal chlorosis | Whiteflies (Bemisia tabaci) | [70] | |
| Tomato yellow leaf curl virus | Common bean, Cowpea | Yellowing and curling of leaves; stunted growth; reduced fruit production; bushy appearance | Whiteflies (Bemisia tabaci) | [71] | |
| Carlavirus | Cowpea mild mottle virus | Cowpea, Soybean, Common bean, Mung bean, Lima bean, Lablab bean | Severe leaf chlorosis, mottling, and distortion; stunting | Whiteflies (Bemisia tabaci); mechanical transmission; seed transmission | [72] |
| Comovirus | Bean pod mottle virus | Soybean | Green to yellow mottling (blotchiness) of young leaves; distortion; stunting; reduced pod size | Bean leaf beetle (Cerotoma trifurcata) | [73] |
| Cowpea severe mosaic virus | Cowpea | Mosaic patterns, leaf deformation, stunting | Bean leaf beetle (Cerotoma arcuata) | [74] | |
| Cucumovirus | Cucumber mosaic virus | Soybean, Peanut, Common bean, Cowpea, Mung bean, Lima bean | Mosaic patterns; leaf distortion; stunting; mottling; chlorosis; necrosis | Aphids (a non-persistent, stylet-borne mechanism); seed transmission | [75] |
| Peanut stunt virus | Peanut, Soybean | Mosaic patterns; vein clearing; leaf rolling; chlorosis; stunting | Aphids | [76] | |
| Cytorhabdovirus | Soybean blotchy mosaic virus | Soybean | Stunting; reduced leaf size; mild mottling; malformed pods; shortening of petioles; leaf crinkling; chlorotic spots | Aphids (Aphis craccivora, A. spiraecola, Myzus persicae) | [77] |
| Emaravirus | Pigeon pea sterility mosaic virus 1 | Pigeon pea | Stunting; bushy growth; reduced leaf size; mosaic patterns on the leaves; excessive vegetative growth; ring spots | Eriophyid mite (Aceria cajani) | [78] |
| Pigeon pea sterility mosaic virus 2 | Pigeon pea | Mosaic patterns on the leaves; stunting; sterility; interveinal chlorosis | Eriophyid mite (Aceria cajani) | [78] | |
| Gammacarmovirus | Soybean yellow mottle mosaic virus | Soybean, Mung bean, Black gram | Leaf mottling; mosaic of light and dark green areas; stunting; reduced pod numbers | Seed transmission | [79] |
| Ilarvirus | Tobacco streak virus | Soybean, Peanut, Mung bean, Black gram | Bud blight; necrotic streaks and rings; leaf distortion; stunting; wilting | Thrips; seed transmission | [80] |
| Luteovirus | Soybean dwarf virus | Soybean | Puckered leaves; interveinal chlorosis; leaf rugosity; stunting | Aphids (Acyrthosiphon solani) | [81] |
| Nanovirus | Faba bean necrotic stunt virus | Common bean | Leaf yellowing; stunting; reddening of leaves; thickening of leaves; suppression of flowering; pod setting | Aphids (Acyrthosiphon pisum, Aphis craccivora, A. fabae) | [82] |
| Milk vetch dwarf virus | Cowpea, Mung bean, Lablab bean | Yellowing; stunting; leaf rolling; crinkling; mosaic; dwarfism | Aphids (Aphis cracciviora) | [83] | |
| Orthotospovirus | Capsicum chlorosis virus | Peanut | Chlorosis; mottling; ringspots; leaf deformation | Thrips (Ceratothripoides claratris, Frankliniella schultzei, Microcephalothrips abdominalis, Thrips palmi) | [84] |
| Groundnut bud necrosis virus | Peanut, Cowpea, Mung bean, Black gram, Lablab bean | Chlorosis; mottling; lesions; stunted growth; necrotic rings; bud necrosis | Thrips (Thrips palmi) | [85] | |
| Groundnut ringspot virus | Peanut, Soybean | Bronzing; mosaic; mosaic with ringspots; yellowing; stem necrosis | Thrips (Frankliniella occidentalis, F. schultzei, F. intonsa, F. gemina) | [86] | |
| Soybean vein necrosis virus | Soybean | Yellowing near leaf veins, eventually turning to reddish-brown lesions | Seed transmission | [87] | |
| Tomato spotted wilt virus | Peanut | Bronzing; curling; necrotic streaks and spots on the leaves; stunting | Thrips (Frankliniella occidentalis); seed transmission | [88] | |
| Potyvirus | Bean common mosaic necrosis virus | Common bean, Lablab bean | Mosaic patterns; necrosis (black root); leaf rolling; blistering; light and dark-green patches; chlorotic vein banding; mottling and malformation of leaves | Aphids (Acyrthosiphon pisum, Aphis fabae, Myzus persicae); seed transmission | [89] |
| Bean common mosaic virus | Common bean, Soybean, Peanut, Cowpea, Mung bean, Black gram, Lablab bean, Bambara groundnut | Mosaic patterns; green vein banding; leaf curling and distortion; stunted growth | Aphids; seeds; pollen | [89] | |
| Bean yellow mosaic virus | Common bean | Mottling; mosaic appearance; leaf distortion; downward cupping; stunting; rough pods | Aphids: mechanical transmission | [90] | |
| Cowpea aphid-borne mosaic virus | Cowpea, Lima bean, Bambara groundnut | Mosaic; mottling; interveinal chlorosis; vein-banding; vein-clearing; vein-yellowing; blistering | Aphids (Aphis craccivora, A. gossypii, A. spiraecola, A. fabae, A. sesbaniae, Macrosiphum euphorbiae, Myzus persicae, Rhopalosiphum maidis, Acyrthosiphon pisum); seed (true seeds) transmission; mechanical transmission | [91] | |
| Peanut mottle virus | Peanut, Soybean | Dark-green mosaic or mottle; crinkled leaflet margins; leaf chlorosis and deformation | Aphids (Aphis craccivora, Aphis gossypii, Hyperomyzus lactucae, Myzus persicae, Rhopalosiphum maidis, Rhopalosiphum padi); peanut seed | [92] | |
| Soybean mosaic virus | Soybean | Vein clearing in the upper trifoliate leaves; downward curling of the leaf margins; raised puffes and puckering; necrosis of the petioles and stems; bud necrosis | Non-specific transmission by aphids; seed transmission | [93] | |
| Sobemovirus | Southern bean mosaic virus | Common bean | Mosaic patterns; leaf distortion | Beetles (Cerotoma trifurcata); seed transmission | [94] |
| Soybean yellow common mosaic virus | Soybean | Leaf mottling, stunting, leaf distortion | Aphids | [95] | |
| Umbravirus | Groundnut rosette virus | Peanut | Chlorotic or green rosette patterns; severe stunting; bushy appearance | Transmitted by Aphis craccivora in the presence of groundnut rosette assistor virus | [96] |
| Unassigned | Groundnut rosette assistor virus | Peanut | Mild mottle symptoms; severe rosette disease symptoms | Transmitted by Aphis craccivora in a persistent manner | [97] |
| Crop | PPN | Pathogen(s) | Reference |
|---|---|---|---|
| Bean | Meloidogyne incognita | Fusarium oxysporum f.sp. phaseoli | [98] |
| Chickpea | M. incognita | F. oxysporum | [99,100] |
| M. javanica | F. oxysporum f.sp. ciceris | ||
| Pratylenchus thornei | |||
| Lentil | M. javanica | F. oxysporum f.sp. lentils | [101] |
| Peanut | Rhizoctonia solani | [102] | |
| Pea | M. incognita Rotylenchulus reniformis | F. oxysporum f.sp. pisi | [103,104] |
| Soybean | Heterodera glycines | F. solani | [105] |
| Phytophthora sojae | [106] |
| Genebank (Location) | Legume Accessions Conserved | Notable Disease-Resistance Contributions |
|---|---|---|
| ICRISAT Genebank (India)—CGIAR Crops: Chickpea, Pigeonpea, Groundnut, etc. | ~20,600 chickpea; ~13,500 pigeonpea; ~15,400 groundnut accessions. (Total ex situ collection > 129,000 accessions across 11 crops) | Germplasm is used worldwide for disease-resistance breeding. For example, ICRISAT provided Fusarium wilt-resistant chickpeas and rust/late leaf spot-resistant groundnuts to national programs. At least 15 chickpea, 10 pigeonpea, and 11 groundnut landraces from this collection were released directly as improved varieties, benefiting farmers with higher yields and disease tolerance. ICRISAT’s mini-core collections have identified multiple resistant sources (e.g., pigeonpea lines resistant to sterility mosaic virus and chickpea to Ascochyta blight). |
| CIAT “Alliance” Bean Collection (Colombia)—CGIAR Crops: Common bean (Phaseolus) and wild relatives | Phaseolus collection ~40,000 accessions (incl. ~36,000 P. vulgaris landraces, plus P. coccineus, P. lunatus, P. acutifolius). Largest global bean collection. | A key source of genetic resistance for bean diseases. CIAT identified rare landraces with broad resistance to angular leaf spot and anthracnose, guiding the breeding of multi-disease-resistant beans. Wild relatives in the collection (e.g., P. coccineus, P. acutifolius) contributed genes for common bacterial blight and bruchid resistance, now incorporated into cultivated bean lines. CIAT-bred “biofortified” high-iron beans also carry stacked resistance to pests and diseases from genebank materials released to small farmers in Africa and Latin America. |
| IITA Genebank (Nigeria)—CGIAR Crops: Cowpea, Soybean, Bambara groundnut, others | >15,000 cowpea (Vigna unguiculata) accessions (world’s largest cowpea collection); also holds >2000 soybean, ~2100 Bambara groundnut, and African yambean, among. Total > 36,000 legume samples. | Enabled the development of multiple disease-resistant cowpea varieties for sub-Saharan Africa. IITA germplasm provided sources for resistance to major cowpea viruses (e.g., CABMV), bacterial blight, and parasitic weed Striga—traits now in improved cultivars grown across West Africa. For instance, the landrace TVu 11986 from IITA’s collection confers broad resistance to Striga and is used in breeding Striga-proof cowpeas. IITA’s pre-breeding with wild Vigna species (like V. vexillata) has yielded lines resistant to cowpea pod borer and fungal diseases, strengthening the crop’s resilience for resource-poor farmers. |
| ICARDA Genebank (Lebanon & Morocco)—CGIAR Crops: Lentil, Chickpea, Faba bean, Pea, Grasspea | ~15,300 chickpea; ~14,370 lentil; ~10,000 faba bean; plus ~4000 grasspea (Lathyrus) and various vetches. It holds ~48% of global faba bean and ~51% of global lentil diversity. Total collection: ~144,000 (including cereals). | Wild relatives and landraces from ICARDA have been pivotal in disease-resistance breeding for cool-season food legumes. Example: lentil accession IG 72815 (a wild Lens ervoides from the ICARDA collection) carries high-level resistance to two races of anthracnose, which has been introduced into cultivated lentil breeding lines. ICARDA’s chickpea landraces from the Mediterranean region provided genes for Ascochyta blight resistance, which is now used in varieties in India and Ethiopia. Faba bean germplasm from ICARDA (e.g., Ethiopian landraces) contributed novel genes for resistance to chocolate spot and faba bean rust in breeding programs. These contributions underscore ICARDA’s role in exploiting West Asian and North African legume diversity for global disease resistance improvement. |
| USDA National Plant Germplasm System (USA) Crops: Soybean, Peanut, Common bean, Pea, etc. | Major US collections: Soybean (Glycine max) ~22,900 accessions; Peanut (Arachis hypogaea) ~9000 accessions; Common bean (Phaseolus vulgaris) ~18,000; Pea (Pisum sativum) ~5500; and thousands of others (lentil, chickpea, forage legumes, wild relatives in genus Glycine, Phaseolus, Arachis, etc). | U.S. collections have yielded critical resistance genes for crop protection. The USDA peanut collection’s PI 203396 (origin: Brazil) famously provided TSWV virus resistance, now widespread in U.S. peanut cultivars. The soybean collection preserves wild Glycine soja and perennial Glycine spp.; from these, genes for soybean rust (Phakopsora) resistance and soybean cyst nematode resistance (e.g., PI 88788 for SCN) have been identified and bred into commercial soybean lines. The common bean collection has contributed sources for BCMV virus resistance and anthracnose (e.g., landrace ‘Jalo’ for rust resistance). Ongoing USDA breeding programs routinely tap these germplasm resources. For example, novel alleles for Fusarium root rot resistance in bean and frog eye leaf spot resistance in soybean have been introgressed from exotic accessions, underscoring the NPGS’s role in safeguarding U.S. crop health. |
| ICAR–NBPGR National Genebank (India) Crops: Diverse grain legumes (pigeonpea, chickpea, mungbean, urd bean, lentils, etc.) | >63,000 legume accessions from 61 species, one of the largest national legume collections. Key holdings: pigeonpea (~12,000), chickpea (~15,000), mungbean (Vigna radiata ~7000), urd bean (V. mungo ~6500), lentil (~3000), lathyrus and others. | Indian legume germplasm has been extensively characterized for disease resistance. NBPGR’s collection underpins breeding fusarium wilt–resistant pulses in India—for example, the pigeonpea variety ‘ICP 8863’ (Maruti) with wilt resistance was derived from a landrace in this genebank. Through multi-institutional evaluation, over 200 Indian germplasm accessions have been registered as donors for important traits, many for biotic stress resistance (e.g., bruchid beetle resistance in rice bean, powdery mildew resistance in pea). NBPGR also facilitated the reintroduction of lost landraces (e.g., a mungbean line with yellow mosaic virus resistance) back into cultivation. This genebank’s trait-diverse collections continue to enhance legume breeding for disease-prone tropical environments. |
| EMBRAPA Genebank Network (Brazil) Crops: Common bean, Soybean, Forages, others | ~18,000 soybean accessions and ~16,000 Phaseolus beans (held at Embrapa Soybean and Embrapa Rice and Beans, respectively). It also conserves Brazil’s cowpea, peanut, and extensive tropical forage legume collections (stylosanthes, brachiaria, etc.). | Embrapa has leveraged its rich germplasm to breed disease-resistant cultivars suited to Brazilian and tropical agriculture. Using the Embrapa collection, the Brazilian common bean program developed the widely grown ‘Carioca’ and ‘Pérola’ bean varieties with multi-disease resistance (to angular leaf spot, anthracnose, rust) derived from landrace crosses. Embrapa’s soybean collection and wild Glycine tomentella accessions enabled the breeding of varieties with improved Asian soybean rust resistance (e.g., the rust-tolerant line BRS 511). Embrapa collaborated with ICRISAT to use wild Arachis introgressions for peanut smut and early leaf spot resistance in peanut. These achievements highlight how Brazil’s national PGR conservation has contributed to more resilient legume crops (and pastures) in the tropics. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilar, C.H.; Pires, D.; Cortaga, C.; Peja, R., Jr.; Cruz, M.A.; Langres, J.; Redillas, M.C.F.; Galvez, L.; Balendres, M.A. Harnessing Legume Productivity in Tropical Farming Systems by Addressing Challenges Posed by Legume Diseases. Nitrogen 2025, 6, 65. https://doi.org/10.3390/nitrogen6030065
Aguilar CH, Pires D, Cortaga C, Peja R Jr., Cruz MA, Langres J, Redillas MCF, Galvez L, Balendres MA. Harnessing Legume Productivity in Tropical Farming Systems by Addressing Challenges Posed by Legume Diseases. Nitrogen. 2025; 6(3):65. https://doi.org/10.3390/nitrogen6030065
Chicago/Turabian StyleAguilar, Catherine Hazel, David Pires, Cris Cortaga, Reynaldo Peja, Jr., Maria Angela Cruz, Joanne Langres, Mark Christian Felipe Redillas, Leny Galvez, and Mark Angelo Balendres. 2025. "Harnessing Legume Productivity in Tropical Farming Systems by Addressing Challenges Posed by Legume Diseases" Nitrogen 6, no. 3: 65. https://doi.org/10.3390/nitrogen6030065
APA StyleAguilar, C. H., Pires, D., Cortaga, C., Peja, R., Jr., Cruz, M. A., Langres, J., Redillas, M. C. F., Galvez, L., & Balendres, M. A. (2025). Harnessing Legume Productivity in Tropical Farming Systems by Addressing Challenges Posed by Legume Diseases. Nitrogen, 6(3), 65. https://doi.org/10.3390/nitrogen6030065







