Abstract
Rhizobia have two major life styles, one as free-living bacteria in the soil, and the other as bacteroids within the root/stem nodules of host legumes where they convert atmospheric nitrogen into ammonia. In the soil, rhizobia have to cope with changing and sometimes stressful environmental conditions, such as nitrogen limitation. In the beta-rhizobial strain Paraburkholderia phymatum STM815, the alternative sigma factor σ54 (or RpoN) has recently been shown to control nitrogenase activity during symbiosis with Phaseolus vulgaris. In this study, we determined P. phymatum’s σ54 regulon under nitrogen-limited free-living conditions. Among the genes significantly downregulated in the absence of σ54, we found a C4-dicarboxylate carrier protein (Bphy_0225), a flagellar biosynthesis cluster (Bphy_2926-64), and one of the two type VI secretion systems (T6SS-b) present in the P. phymatum STM815 genome (Bphy_5978-97). A defined σ54 mutant was unable to grow on C4 dicarboxylates as sole carbon source and was less motile compared to the wild-type strain. Both defects could be complemented by introducing rpoN in trans. Using promoter reporter gene fusions, we also confirmed that the expression of the T6SS-b cluster is regulated by σ54. Accordingly, we show that σ54 affects in vitro competitiveness of P. phymatum STM815 against Paraburkholderia diazotrophica.
Keywords:
rhizobia; σ factor; RpoN; RNA-Sequencing; nitrogen; motility; type VI secretion system (T6SS) 1. Introduction
Symbioses between legumes and rhizobia increase soil fertility and crop yield by means of biological nitrogen fixation [1]. Rhizobia are soil bacteria, which adapt to different environmental stresses and eventually nodulate the roots or the stems of compatible legume plants. In the symbiotic organ—the nodule—bacteria live inside the host plant cells and eventually differentiate into nitrogen-fixing bacteroids. Bacteroids convert atmospheric nitrogen (N2) into ammonia that is assimilated and used as a nitrogen source by the plant [2,3]. To support this energetically expensive reaction (16 molecules of ATP and 8 low potential electrons per N2 reduced), the legume provides the bacteroids with energy in form of reduced carbon compounds. C4-dicarboxylates such as succinate, fumarate, and malate have been shown to be the primary carbon source used by bacteroids and are oxidized to CO2 in the tricarboxylic acid cycle [4].
Rhizobia are phylogenetically diverse and belong to the alpha-proteobacterial (alpha-rhizobia) and the beta-proteobacterial group (beta-rhizobia). Beta-rhizobial strains such as Burkholderia and Cupriavidus were first isolated from nodules in 2001 [5,6], and most of the legume nodulating Burkholderia strains were recently re-classified into the new Paraburkholderia genus [7,8]. Symbiotic Paraburkholderia species have been mainly isolated from nodules of Mimosa plants in South America and South East Asia [9,10,11,12], but also from South African Fynbos [13,14,15,16]. Geographical position, environment, host plant, and coevolution with the symbiont have been shown to affect the presence and dominance of certain rhizobial species in soil and in nodules [17,18]. Paraburkholderia phymatum STM815 is an interesting strain since it is able to nodulate mimosoid as well as papilionoid legumes. Furthermore, it has been shown by several groups to be highly competitive in infecting the roots of mimosoid and papilionoid legumes [17,19,20,21]. A comparison of the phenotypic traits of several Paraburkholderia strains showed that P. phymatum STM815 produces large amounts of exopolysaccharides (EPS), is very motile, and able to outcompete other Paraburkholderia strains in vitro [20]. Our group has also shown that P. phymatum STM815 harbors two type VI secretion systems (T6SS) in its genome, which contribute to the competitive ability of this strain in vitro and in infecting plants [22]. These characteristics partly explain the success of this strain in competing with other rhizobia in the soil. However, the regulatory networks underlying the high competitiveness of P. phymatum STM815 in infecting several legumes are still unknown.
The σ factor σ54 is structurally distinct from the σ70-type sigma factors, recognizes different promoter elements located at position −24 and −12 upstream of the transcription start site [23], and requires an enhancer-binding protein (EBP) to activate transcription of target genes. Usually bacteria encode multiple different EBPs in their genome, with each of them controlling different traits required to adapt to specific ecological niches [24]. We recently showed that the alternative σ factor σ54 is a key regulator of P. phymatum STM815 symbiotic nitrogen fixation inside root nodules of Phaseolus vulgaris (common bean) [25]. In fact, a σ54 mutant did not form an efficient symbiosis with P. vulgaris and was impaired in reducing N2 to ammonium. By using RNA-Seq and metabolomics on bean root nodules formed by P. phymatum STM815 wild type and a σ54 mutant [26], we found that in addition to the symbiotic genes, σ54 also controls several genes potentially important for P. phymatum STM815 to persist in soil.
In this study, we analyzed the regulon of σ54 (Bphy_0326) by growing the cells under free-living conditions in a nitrogen-limited medium. Among the top genes activated by σ54, we found dctA, which codes for a C4 dicarboxylate transporter, a flagellar gene cluster, and one of the two T6SS present in the genome of P. phymatum STM815 (T6SS-b). Indeed, we confirmed that the σ54 mutant was not able to grow on C4 dicarboxylates, was impaired in swimming motility, and that both these defects were complemented by providing rpoN in trans. Moreover, the mutation in rpoN affected the expression of the T6SS-b gene cluster and rendered P. phymatum STM815 slightly less competitive against Paraburkholderia diazotrophica [27], suggesting that σ54 is involved in the control of interbacterial competition.
2. Materials and Methods
2.1. Bacterial Strains, Media, and Cultivation
The strains, plasmids, and primers employed in this study are listed in Table S1. Escherichia coli cells were routinely grown in Luria-Bertani liquid medium (LB), whereas P. phymatum STM815 strains were cultivated in LB salt-free liquid medium [20].
The bacterial cultures used for RNA-Seq were prepared by growing P. phymatum cells in a modified AB minimal medium [28], with 10 mM sodium citrate (Sigma-Aldrich) as carbon source. The nitrogen source in AB minimal medium (NH4)2SO4 was replaced with either 30 mM NH4Cl (Sigma-Aldrich, to obtain nitrogen-replete conditions) or 0.5 mM NH4Cl (nitrogen-limited condition). Na2SO4 (Sigma-Aldrich) was added to AB minimal medium to obtain a final concentration of 15 mM of sulfate. Bacterial cultures were grown in 250 mL Erlenmeyer flasks containing 100 mL medium and incubated on a shaker (220 rpm) at 30 °C. P. phymatum STM815 wild-type and σ54 mutant cells were grown to exponential phase (OD600 = 0.4–0.5) in minimal medium under nitrogen-replete conditions, then washed twice in AB minimal medium without a nitrogen source and incubated further aerobically for one hour under nitrogen-limited conditions. For each strain, three independent biological replicates were prepared and processed for RNA-Seq analysis.
To test the induction of the promoter fusions, the bacteria were grown at 30 °C in AB medium containing 10 mM glucose as C-source (ABG) in a 96-well plate (Falcon, Corning, USA) to exponential phase. The optical density and GFP fluorescence (excitation/emission wavelength equal to 488 nm/520 nm) were measured using a TECAN plate reader (TECAN Infinite M200 PRO, Tecan Trading AG, Switzerland).
The growth of the following P. phymatum STM815 strains—wild type (wt), σ54 mutant, and σ54 complemented—was tested in AB minimal medium with 15 mM of three different C4-carbon sources: Fumaric acid (Sigma-Aldrich, ABF), malic acid (Sigma-Aldrich, ABM), and succinic acid (Sigma-Aldrich, ABS). For each strain and C-source tested, the growth of three independent biological replicates was measured.
2.2. Promoter Fusion Construction
To construct promoter fusions, the two promoters of interest were introduced into the vector pPROBE-NT (Table S1) [29]. To construct pPROBE-T6SS-b (p5978), the promotor of T6SS-b was amplified from P. phymatum STM815 genomic DNA (gDNA) with the primers Bphy_5978_Promotor_F_HindIII and Bphy_5978_Promotor_R_BamH1 (Table S1). The 657 bp long PCR product was restricted with the enzymes HindIII and BamHI and cloned into pPROBE-NT. The correct sequence was confirmed by sequencing at Microsynth (Balgach, Switzerland) using pCO13-R3 primer [30]. The plasmid was then conjugated into P. phymatum STM815 wild type and σ54 mutant, and the transconjugants were selected on ABS plates. To obtain the second promoter fusion, the promotor region of the T6SS-3 cluster (p6115) was amplified using primers p6115_SalI_For and p6115_EcoRI_rev (Table S1). The obtained PCR fragment (480 bp) was digested and cloned into pPROBE-NT between the SalI and EcoRI sites. Once the sequence of the construct had been confirmed through sequencing at Microsynth using pCO13-R3 primer [30], the plasmid was mobilized into P. phymatum STM815 wild type and σ54 mutant.
2.3. RNA-Sequencing and Data Processing
Total RNA was isolated from flash-frozen, pelleted cells of the wild type and the σ54 mutant using a modified hot acid phenol protocol [31]. Afterwards, gDNA was digested by DNase treatment and the quality of the total RNA as well as the complete removal of gDNA were checked by PCR [25,26]. The cDNA synthesis was initiated with 150 ng of total RNA and the library was prepared and purified with the Ovation® Complete Prokaryotic RNA-Seq DR Multiplex Systems (NuGEN, San Carlos, CA, USA) [25,26]. Next, the quality and quantity of the cDNA libraries were analyzed with a TapeStation System (Agilent Technologies, Santa Clara, CA, USA). The prepared cDNA libraries were single-end sequenced with a HiSeq2500 instrument (Illumina, San Diego, CA, USA) at the Functional Genomic Center Zurich (FGCZ). Using the CLC Genomics Workbench v7.0 (QIAGEN CLC bio, Aarhus, Denmark) program, the obtained reads were trimmed to 70 bp and mapped to the P. phymatum STM815 genome, allowing up to two mismatches per read [32]. Afterwards the unique reads were analyzed statistically with the DESeq R-package (version 1.30.0) [33]. The top 200 significantly RpoN-regulated genes with a log2(FC) ≥ 1 and ≤ −1 were considered and then ranked by ascending p-value. To get additional information, the top 200 differentially regulated genes were assigned to functional categories (EggNOG v3.0) [34]. Table S2 lists all P. phymatum STM815 genes and their expression profile in the wild type and the σ54 mutant. The RNA-Seq raw and processed data files of the P. phymatum STM815 wild-type and σ54 mutant strains are available with the GSE156048 accession number.
2.4. Phenotypic Analysis
Plates for the swimming motility test were prepared by using LB salt-free medium containing 1% tryptone (Difco), 0.5% yeast extract (Difco), and 0.2% agar. The bacterial cells were washed twice with 10 mM MgSO4 and normalized to an OD600 of 0.5. The plates were inoculated with the bacterial suspension using a 20 μL pipette tip. Subsequently, the plates were incubated at 30 °C inside a box containing wet paper towels. The diameter of the swimming zone was measured after 40 h incubation.
To perform competition experiments on plates, a target strain and an attacker strain were chosen, plated on salt-free LB without antibiotics and incubated overnight at 30 °C. The strains were washed twice with 10 mM MgSO4 and the OD600 was adjusted (target strain OD600 = 0.2, attacker strain OD600 = 2.0). The strains were mixed in a 1:1 volume ratio. Twenty µl aliquots of the mixture were put on cellulose nitrate membrane filters (GE Healthcare Life Sciences, Marlborough, MA, USA) on ABG plates, and incubated at 30 °C. After 24 h, the cells on the filters were resuspended in one ml 10 mM MgSO4. The colony forming units (CFU) were determined on salt-free LB plates, both with and without antibiotics, to recover the attacker and target strains, respectively.
2.5. Statistical Analysis
For category distribution, the percentages of upregulated and downregulated genes in each category were calculated as described previously [25]. One-way ANOVA (performed with Prism 7.0), in which the mean of each strain was compared to the mean of the wild type, was used to analyze the motility data. The one unpaired t-test corrected for multiple comparisons using the Holm–Sidak method (Prism 7.0) was employed to statistically analyze the results of the competition on plate. The induction of the promoter fusions was tested with two-way ANOVA corrected for multiple comparisons using the Sidak test (Prism 7.0).
3. Results
3.1. The P. phymatum STM815 σ54 Regulon in Free-Living Nitrogen-Limiting Conditions
RNA-Seq was employed to determine the P. phymatum STM815 σ54 regulon under nitrogen-limited free-living conditions. For each strain, three independent biological replicates were prepared, processed, and analyzed as described previously [25]. The unique read counts obtained per sample ranged from 6.9 to 13.2 million reads.
After a DESeq analysis comparing gene expression in the P. phymatum STM815 σ54 mutant versus wild type, the top 200 differentially regulated genes (DESeq analysis p-value < 2.2 × 10−8, with log2 [FC] ≥ 1 and ≤ −1) were identified and classified in eggNOG functional categories [34] (Figure 1). Among these 200 top-regulated genes, 81 showed increased expression, while 119 were downregulated in the σ54 mutant (and were therefore positively regulated by σ54). While the two categories “signal transduction mechanisms” and “cell motility” were over-represented among the downregulated genes (Table 1), one category (“cell wall, membrane, envelope biogenesis”) was found to be over-represented in the upregulated genes.
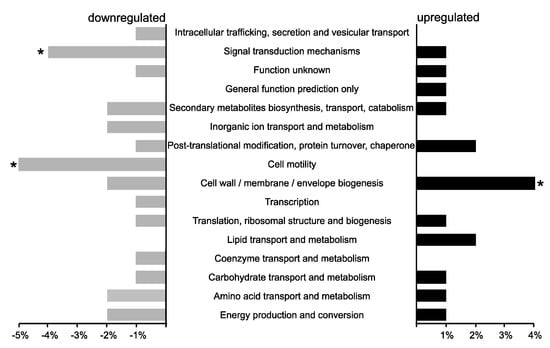
Figure 1.
Functional categories of the top 200 differentially expressed genes of the P. phymatum STM815 wild type versus σ54 mutant (gray, downregulated genes, black upregulated genes) under nitrogen starvation according to eggNOG classification [34]. The asterisks (*) indicate statistical significance for over-represented genes in a particular category (p-value < 0.02).

Table 1.
List of 119 genes positively controlled by σ54. Over-represented categories are marked with and asterisk (Fischer test, p-value < 0.02). Genes harboring a putative RpoN-box in their promoter region are shown in bold.
In the category “cell motility”, we found several genes involved in flagella biosynthesis (in the Bphy_2926-64 cluster) and a gene involved in chemotaxis (Bphy_5592). In the category “signal transduction mechanisms”, several transcriptional regulatory genes (e.g., Bphy_3958, Bphy_5314, and Bphy_5669) and genes coding for histidine kinases (e.g., Bphy_0226, Bphy_3957, Bphy_3963, Bphy_4749, Bphy_5668, and Bphy_5975) were identified. Among these top differentially regulated genes, six might be part of a two-component regulatory system (TCRS): Bphy_3957-58, Bphy_3962-63, and Bphy_5668-69. Upstream of Bphy_5668-69, we found Bphy_5667, which codes for a citrate carrier and has an RpoN-binding sequence in the promoter region (Figure 2A). Moreover, downstream of Bphy_5668-69 we found a gene coding for a diguanylate phosphodiesterase potentially involved in c-di-GMP metabolism (Bphy_5670, Figure 2A). The histidine kinase Bphy_0226 (dctB) is located downstream of a gene coding for a C4-dicarboxylate transporter (dctA, Bphy_0225), which was also on the list of top-regulated genes and contained an RpoN box in the promoter region (Figure 2B). In several bacteria, the DctA transporter has been shown to be involved in the assimilation of C4 dicarboxylates, i.e., fumarate, malate, and succinate [4,35,36], which are important energy sources for symbiotic rhizobia [37]. Interestingly, most of the genes (13 of 20) belonging to one of the two T6SS clusters present in P. phymatum STM815 (Bphy_5978-97, T6SS-b, Figure 2C) grouped in the category “function unknown” were downregulated in the σ54 mutant grown under nitrogen starvation. By contrast, the expression of the second T6SS cluster (Bphy_6107-6129, T6SS-3) was not dependent on σ54. T6SSs are contact-dependent secretion systems that have been shown to be important for the transport of effectors to other prokaryotic or eukaryotic cells [38]. Interestingly, several genes located in a cluster coding for components of a type IV secretion system (T4SS, Bphy_7524-37) also showed significant downregulation in the σ54 mutant. Similar T4SSs are usually involved in conjugation [39].
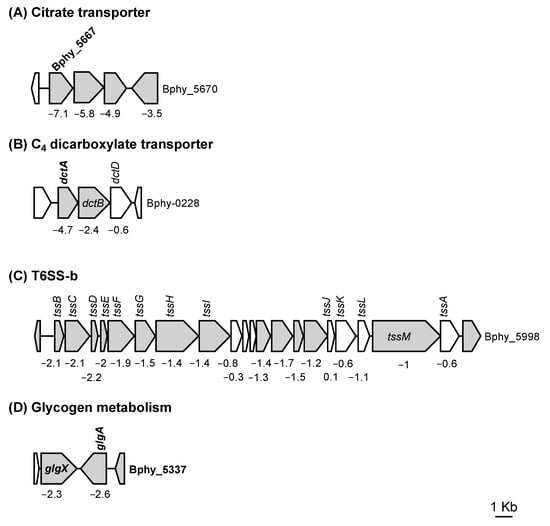
Figure 2.
Four P. phymatum STM815 gene clusters showing significant downregulation in a σ54 mutant compared to the wild type: (A) Citrate carrier protein and associated two-component regulatory system (TCRS), (B) C4 dicarboxylate transporter T6SS-b, (C) T6SS-b, and (D) glycogen metabolism. Genes containing an RpoN box in their promoter region are indicated in bold. The top 200 regulated genes are colored gray. The log2 of the fold change has been indicated below the genes in the cluster of interest.
In the category “carbohydrate transport and metabolism”, we found several genes whose expression was downregulated in the σ54 mutant: Bphy_2145, coding for a trehalose-6-phosphate synthase (otsA), as well as two genes involved in glycogen metabolism (Bphy_5335 and Bphy_5336, glgX and glgA, Figure 2D). All three genes display an RpoN-binding sequence in their promoter region (Table 1). In the operon Bphy_3959-61, which encodes a potential nitrate/sulfonate/bicarbonate transporter, we found the most highly downregulated genes (log2(FC) = −7.1, −7.4, −6.0, for Bphy_3959 to Bphy_3961, respectively). Additionally, the expression of three genes in a cluster potentially coding for a polysaccharide (Bphy_3730-1 and Bphy_3733) was downregulated in the σ54 mutant compared to the wild type.
As previously mentioned, among the 81 genes upregulated in the mutant, the category “cell wall/membrane/envelope biogenesis” was over-represented, with several genes coding for porins (Bphy_0154, Bphy_1082 and Bphy_2684), as well as a cluster coding for the polysaccharide cepacian (Bphy_1056-77) [40]. Out of the 200 genes significantly regulated by RpoN, ten were located on the symbiotic plasmid including the T4SS and an aminotransferase class-III (Bphy_7651), which were all positively regulated by σ54.
By applying a less stringent p-value cut-off than that selected for the top 200 differentially regulated genes (DESeq analysis p-value ≤ 2 × 10−5), additional genes associated with nitrogen metabolism were found to be positively regulated by σ54, such as the TCRS ntrBC (Bphy_1479-80), urtB (Bphy_2252) coding for a urea ABC transporter permease and the glnB1-amtB operon, coding for the nitrogen regulatory protein P-II and the ammonium transporter (Bphy_0256-57).
3.2. Phenotypic Characterization of the σ54 Mutant
3.2.1. Role of σ54 for Assimilation of C4 Dicarboxylates
As mentioned above, one of the genes significantly downregulated under nitrogen-limited conditions in our transcriptomic analysis was dctA (Bphy_0225), which codes for a C4 dicarboxylate transporter. DctA is located upstream of a gene cluster encoding the sensor kinase DctB and the response regulator DctD, which belongs to the σ54-interacting protein family. While dctB expression was downregulated in absence of σ54, dctD expression did not change, suggesting that dctD is in a separate transcriptional unit (Figure 2B). In the promoter region of dctA, an RpoN-binding box was identified.
A similar genomic constellation to dctA-dctB-dctD is found for dicarboxylate transport systems in several bacteria, however, dctB and dctD are usually transcribed divergently compared to dctA [4]. To verify that expression of the DctA transporter gene was regulated by σ54, we examined the ability of the wild-type, the σ54 mutant, and the complemented σ54 strains to grow in minimal medium in the presence of C4-carbon sources. The growth of these P. phymatum STM815 strains was tested in defined buffered AB minimal medium supplemented with fumaric acid, malic acid, and succinic acid. We found that the σ54 mutant strain was impaired in the utilization of all tested C4 organic acids and that this defect could be restored in the σ54 complemented strain (Table 2). The results confirmed the validity of our transcriptomic analysis and suggested that σ54 is involved in the assimilation of C4 compounds in P. phymatum STM815.

Table 2.
Utilization of different C4 sources by P. phymatum STM815 wild-type (wt), σ54 mutant (σ54 mt), and σ54 complemented (σ54 comp) strains 1.
3.2.2. σ54 Positively Controls Swimming Motility
In the list of top genes positively regulated by σ54 at the transcript level (Table 1), the category “cell motility” was over-represented. The expression of Bphy_2938-39 (fliMN), Bphy_2956 (flgJ), and Bphy_2962 (flhF), all of which encode flagellar biosynthesis proteins, was significantly downregulated in a σ54 mutant (Table 1). Since flagella are used for motility, the swimming ability of P. phymatum STM815 wild type, the σ54 mutant, and the complemented strain was tested on salt-free LB plates, and the diameter of the swimming cells was measured after 40 h of incubation at 30 °C. In agreement with our RNA-Seq data, the σ54 mutant was 40% less motile, and the complemented strain showed increased motility relative to the wild-type strain (Figure 3).
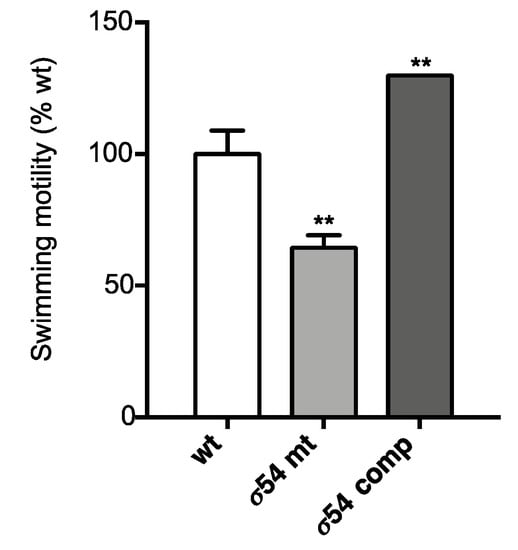
Figure 3.
Swimming motility of P. phymatum STM815 wild-type (wt), σ54 mutant strain (σ54 mt) and σ54 mutant complemented strain (σ54 comp) on salt-free Luria-Bertani LB plates. The plates were incubated at 30 °C and the diameter measured after 40 h. The experiment was performed in triplicates, and the obtained results were analyzed using one-way ANOVA (**, p-value ≤ 0.01). The standard deviation is shown.
3.2.3. The Presence of σ54 Influences Interbacterial Competition
Our RNA-Seq data showed that the expression of one P. phymatum STM815 cluster coding for a T6SS (Bphy_5978-97, T6SS-b cluster) was dependent on σ54 under nitrogen-limiting growth conditions (Figure 2C). Both P. phymatum STM815 T6SS systems were previously shown to be important for competition with other related Paraburkholderia strains such as P. diazotrophica [22]. In order to validate the RNA-Seq data, the promoters of Bphy_5978 (T6SS-b cluster) or Bphy_6115 (T6SS-3 cluster) were fused with gfp, and expression was measured in the P. phymatum STM815 wild-type and the σ54 mutant background. In accordance with the transcriptome data, only the expression of the T6SS-b cluster (p5978), and not of the T6SS-3 cluster (p6115), was decreased in a P. phymatum STM815 σ54 mutant compared to the wild-type strain (Figure 4).
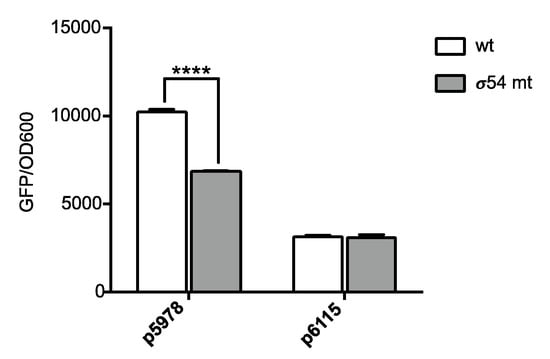
Figure 4.
Expression of the T6SS-b cluster (p5978) and the T6SS-3 cluster (p6115) in P. phymatum STM815 wild-type (wt) and σ54 mutant (σ54 mt) strains. The histograms show gfp activity from the promoter (measured at a wavelength of 488 nm) normalized by the OD600. Three independent biological replicates were performed. The statistical analysis was performed using two-way ANOVA corrected with Sidak’s multiple comparisons test. ****, p < 0.0001.
We next tested the ability of the P. phymatum STM815 wild-type, the σ54 mutant, and the complemented strain to compete against P. diazotrophica for 24 h on ABG minimal medium (Figure 5). The σ54 mutant turned out to be slightly, but significantly, less competitive as compared to the P. phymatum STM815 wild type, indicating a role for this alternative sigma factor in controlling interbacterial competition. In line with this observation, a σ54 complemented strain, which expressed rpoN from the the lac promoter on the pBBR1MCS plasmid, out-competed P. diazotrophica more effectively than the wild-type strain.
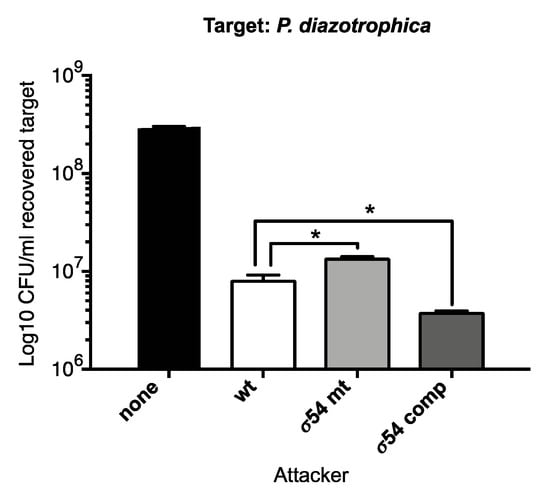
Figure 5.
Competition assay with P. phymatum STM815 wild-type (wt), σ54 mutant (σ54 mt), and σ54 complemented (σ54 comp) strains using P. diazotrophica as target. The assay was performed on ABG plates for 24 h at 30 °C. The histogram shows the mean colony forming units (CFU) per ml of the recovered P. diazotrophica strain (target). Three independent biological replicates were carried out, and error bars indicate the standard deviation. The results were analyzed using an unpaired t-test corrected for multiple comparisons using the Holm-Sidak test. *, p < 0.01.
4. Discussion
The alternative sigma factor σ54 was originally discovered in Salmonella as the sigma factor required for the synthesis of glutamine synthetase [41]. Subsequently, σ54 was identified in a wide range of Gram-negative and Gram-positive bacteria, and found to play a general role in the control of nitrogen metabolism and to affect various other cellular functions such as motility, dicarboxylate transport, degradation of xenobiotics, and virulence in plant and human pathogens [42,43,44,45,46,47,48].
In the rhizobia, including P. phymatum STM815, σ54 is a master regulator during symbiosis since it controls the expression of the genes encoding nitrogenase, i.e., the enzyme that converts N2 into a form that can be readily assimilated by the plant [49,50].
P. phymatum STM815 is a beta-rhizobium that previously belonged to the genus Burkholderia, which comprises versatile bacteria able to adapt and colonize different environmental niches. Our group has shown that in the closely related opportunistic pathogen Burkholderia cenocepacia H111, σ54 is an important regulator of several phenotypic traits, such as the utilization of nitrogen sources, motility, EPS and biofilm formation, biosynthesis of poly-hydroxybutyrate (PHB), and virulence towards Caenorhabditis elegans [48]. To control biofilm formation in H111, σ54 interacts with the EBP BerB, which binds c-di-GMP and thereby regulates expression of the exopolysaccharide Bep [51].
We report here that σ54 also plays an important role in P. phymatum STM815 grown under free-living conditions, where it regulates motility and the expression of T6SS-b, and controls the uptake of tricarbocylic acid cycle TCA cycle intermediates such as succinate, fumarate, and malate (Figure 6). Depending on the presence or absence of oxygen in the environment, dicarboxylates are taken up by bacteria using transporters of different protein families. The dicarboxylate transporter DctA, which belongs to the dicarboxylate amino acid-cation symporter (DAACS) family, has been shown to facilitate dicarboxylate uptake under aerobic conditions in several bacteria, including E. coli and Rhizobium leguminosarum [4,52,53]. We demonstrate here that the P. phymatum STM815 σ54 mutant is unable to take up succinate, fumarate, and malate under aerobic, free-living conditions, and that the expression of dctA (Bphy_0225), which is located upstream of the two component regulatory system genes dctBD, is activated by σ54. DctB is a sensor kinase located in the membrane, which is autophosphorylated in the presence of C4 dicarboxylates and transfers a phosphate group to its cognate response regulator DctD. DctD is a σ54-dependent EBP that activates transcription of target genes including dctA [35]. DctA has been shown to be essential for symbiotic nitrogen fixation in Sinorhizobium meliloti and R. leguminosarum [54,55,56]. We are currently evaluating whether dctA is the main dicarboxylate transporter in P. phymatum STM815 and its role during symbiosis. Interestingly, the expression of the B. cenocepacia H111 dctA ortholog (I35_RS14715) is not regulated by σ54, suggesting that C4 uptake may not be dependent on this alternative sigma factor in opportunistic pathogens of the genus Burkholderia.
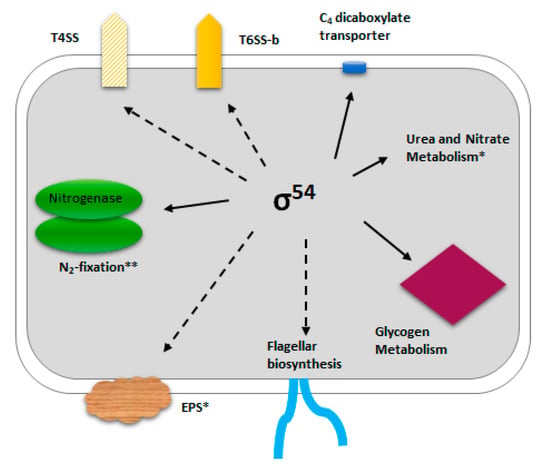
Figure 6.
Schematic representation of the phenotypic traits regulated by σ54 in P. phymatum STM815. Direct σ54 targets (with an RpoN-binding motif in the promoter region) are indicated with a solid line. * Lardi et al. 2017; ** Lardi et al. 2018; T6SS, Type 6 Secretion System; T4SS, Type 4 Secretion System; EPS, Exopolysaccharide.
Among the most highly downregulated P. phymatum STM815 genes (Table 1), we found Bphy_3959-61, which encodes a potential nitrate/sulfonate/bicarbonate ABC transporter. Out of the five genes annotated as citrate transporter in the P. phymatum STM815 genome (Bphy_0810, Bphy_3035, Bphy_4278, Bphy_5667, and Bphy_5880), only Bphy_5667 was significantly regulated by σ54 (Figure 2A), suggesting that this transporter plays a key role in importing citrate under free-living conditions.
Several genes potentially involved in glycogen and trehalose metabolism were found among those that were statistically most significantly regulated by σ54. In fact, the expression of genes encoding a glycogen synthase (Bphy_5336, glgA) (Figure 2D), a glycogen debranching enzyme (Bphy_5335, glgX) which degrades glycogen, and a trehalose-6-phosphate synthase (Bphy_2145, otsA) was activated by σ54 and RpoN-binding motifs were found in the promoter regions of these genes (Table 2).
Glycogen is a soluble polysaccharide composed of glucose in an α-1,4-linked linear arrangement with α-1,6-branches that serves as a storage molecule in many organisms, including eukaryotes and prokaryotes. During starvation periods, glycogen provides a source of stored energy and carbon. The non-reducing disaccharide trehalose (composed of two molecules of α –D-glucose) also acts as an energy reserve compound as well as an osmoprotectant. In Bradyrhizobium diazoefficiens, trehalose is produced to allow survival during desiccation, oxidative stress, and during nodule senescence [57,58,59,60,61,62]. The precursor of trehalose, trehalose 6-phosphate, has been proposed to be a signal for plant growth and stress tolerance in Rhizobium etli [63]. In plants, trehalose- 6-phosphate is an important signal metabolite that coordinates carbon and nitrogen metabolism [64,65,66]. The three genes (glgA, glgX and otsA) are regulated by σ54 not only in free-living conditions, but also during symbiotic growth in root nodules [26]. Additionally, the gene encoding a trehalose synthase (Bphy_7407) was regulated by σ54 during symbiosis, but not under free-living conditions. To the best of our knowledge, the regulation of glycogen and trehalose synthesis via σ54 has not been reported previously. Trehalose biosynthesis was previously shown to be regulated by sigma factor σ38 (RpoS) in E. coli [67] and by the extra-cytoplasmic function (ECF) sigma factor RpoE2 of S. meliloti [68]. Interestingly and in contrast to H111, the synthesis of the storage compound PHB is not controlled by σ54 in P. phymatum STM815.
To date, not much is known about the regulation of T6SS expression in rhizobia. In this study, we show that σ54 is involved in the regulation of one of the two P. phymatum STM815 T6SS gene clusters (Figure 2 and Figure 4), and that a σ54 mutant strain is slightly less competitive with P. diazotrophica than wild type P. phymatum STM815 (Figure 5). RpoN regulation of T6SS-b is probably indirect, since we did not find a RpoN-binding sequence in the promoter region of the first gene in the operon. However, the presence of two dicarboxylate transporter genes in T6SS-b (Bphy_5990 and Bphy_5992) could be the reason for the indirect control of this cluster by σ54. Our results also suggest that the two P. phymatum STM815 T6SSs are used under different conditions and are therefore subject to different regulatory mechanisms, with T6SS-3 expression being σ54-independent. Bernard and colleagues [69] showed that σ54 controls expression of T6SSs in several environmental strains, including Vibrio cholerae, Aeromonas hydrophila, Marinomonas MWYL1 and the plant pathogenic bacteria Pectobacterium atrosepticum, and Pseudomonas syringae pv. tomato [69]. Interestingly, several T6SS clusters investigated to date contain an EBP encoding gene (vasH), which has been demonstrated to be required for maximal activation of T6SS expression [69,70]. No vasH homolog was apparent in the two T6SS clusters present in P. phymatum STM815. Additionally, chromatin immunoprecipitation coupled with next-generation sequencing (ChIP-Seq) and RNA-Seq experiments performed in V. cholerae identified two genes coding for two T6SS hallmark proteins, Hcp and VgrG3, as direct σ54 targets [71,72].
In summary, in the beta-rhizobial strain P. phymatum STM815 σ54 controls important phenotypic traits, such as motility, competition, and transport of C4-dicarboxylates, which all contribute to the high competitiveness of this strain in infecting the roots of various legumes. The identity of the different EBPs that work together with σ54 to control expression of these different traits will be an interesting area for future investigations.
Supplementary Materials
The following are available online at https://www.mdpi.com/2504-3129/1/2/8/s1, Table S1: Bacterial strains and plasmids used in this study, Table S2: List of all P. phymatum STM815 genes, log2 of the fold changes in expression and the p-values in free-living conditions under nitrogen starvation by σ54 mutant versus wild type as assessed by DESeq analysis.
Author Contributions
M.L. and G.P. conceived and designed experiments; M.L., Y.L., S.H., and S.B.d.C. performed the experiments; M.L., Y.L., S.H., L.E., and G.P. analyzed the data; M.L. and G.P. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Swiss National Science Foundation grant number 31003A_179322 to Gabriella Pessi.
Acknowledgments
We are thankful to Christian Ahrens (Agroscope, Switzerland) for critical feedback on the manuscript and Kirsty Agnoli-Antkowiak for careful proofreading. We thank Catherine Aquino Fournier and Lennart Opitz from the Functional Genomics Center Zurich (FGCZ) for support in RNA-Seq data generation.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Vitousek, P.M.; Hättenschwiler, S.; Olander, L.P.; Allison, S. Nitrogen and Nature. Ambio 2002, 31, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.; Kahn, D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, J.J.; Hood, G.A.; Poole, P.S. What determines the efficiency of N2-fixing Rhizobium-legume symbioses? Adv. Microb. Physiol. 2012, 60, 325–389. [Google Scholar] [CrossRef]
- Yurgel, S.N.; Kahn, M.L. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 2004, 28, 489–501. [Google Scholar] [CrossRef]
- Moulin, L.; Munive, A.; Dreyfus, B.; Boivin-Masson, C.; Munive, J.-A. Nodulation of legumes by members of the β-subclass of Proteobacteria. Nature 2001, 411, 948–950. [Google Scholar] [CrossRef]
- Chen, W.M.; Laevens, S.; Lee, T.M.; Coenye, T.; De Vos, P.; Mergeay, M.; Vandamme, P. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 2001, 51, 1729–1735. [Google Scholar] [CrossRef]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef]
- Beukes, C.W.; Palmer, M.; Manyaka, P.; Chan, W.Y.; Avontuur, J.R.; Van Zyl, E.; Huntemann, M.; Clum, A.; Pillay, M.; Palaniappan, K.; et al. Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 2017, 8, 1154. [Google Scholar] [CrossRef]
- Chen, W.-M.; De Faria, S.M.; Straliotto, R.; Pitard, R.M.; Simões-Araújo, J.L.; Chou, J.-H.; Chou, Y.-J.; Barrios, E.; Prescott, A.R.; Elliott, G.N.; et al. Proof that Burkholderia strains form effective symbioses with legumes: A study of novel Mimosa-nodulating strains from South America. Appl. Environ. Microbiol. 2005, 71, 7461–7471. [Google Scholar] [CrossRef]
- Liu, X.; Wei, S.; Wang, F.; James, E.; Guo, X.; Zagar, C.; Xia, L.G.; Dong, X.; Wang, Y.P. Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in Southern China. FEMS Microbiol. Ecol. 2012, 80, 417–426. [Google Scholar] [CrossRef]
- Mishra, R.P.; Tisseyre, P.; Melkonian, R.; Miché, L.; Klonowska, A.; Gonzalez, S.; Laguerre, G.; Chaintreuil, C.; Béna, G.; Moulin, L. Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: Investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol. Ecol. 2012, 79, 487–503. [Google Scholar] [CrossRef] [PubMed]
- Gehlot, H.S.; Tak, N.; Kaushik, M.; Mitra, S.; Chen, W.-M.; Poweleit, N.; Panwar, D.; Poonar, N.; Parihar, R.; Tak, A.; et al. An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann. Bot. 2013, 112, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, B.; Dlodlo, O.; Chimphango, S.; Stirton, C.; Schrire, B.; Boatwright, J.S.; Honnay, O.; Smets, E.; Sprent, J.; James, E.K.; et al. Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa). FEMS Microbiol. Ecol. 2015, 91, 1–17. [Google Scholar] [CrossRef]
- Lemaire, B.; Chimphango, S.B.M.; Stirton, C.; Rafudeen, M.S.; Honnay, O.; Smets, E.; Chen, W.-M.; Sprent, J.; James, E.; Muasya, A.M. Biogeographical patterns of legume-Nodulating Burkholderia spp.: From African fynbos to continental scales. Appl. Environ. Microbiol. 2016, 82, 5099–5115. [Google Scholar] [CrossRef]
- Gyaneshwar, P.; Hirsch, A.M.; Moulin, L.; Chen, W.-M.; Elliott, G.N.; Bontemps, C.; Estrada-de los Santos, P.; Gross, E.; Dos Reis, F.B.; Sprent, J.I.; et al. Legume-nodulating Betaproteobacteria: Diversity, host range, and future prospects. Mol. Plant Microbe Interact. 2011, 24, 1276–1288. [Google Scholar] [CrossRef]
- Howieson, J.; De Meyer, S.E.; Vivas-Marfisi, A.; Ratnayake, S.; Ardley, J.K.; Yates, R. Novel Burkholderia bacteria isolated from Lebeckia ambigua—A perennial suffrutescent legume of the fynbos. Soil Biol. Biochem. 2013, 60, 55–64. [Google Scholar] [CrossRef]
- Melkonian, R.; Moulin, L.; Béna, G.; Tisseyre, P.; Chaintreuil, C.; Heulin, K.; Rezkallah, N.; Klonowska, A.; Gonzalez, S.; Simon, M.F.; et al. The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype: Competition for nodulation in α- and β-rhizobia. Environ. Microbiol. 2014, 16, 2099–2111. [Google Scholar] [CrossRef]
- Bontemps, C.; Rogel, M.A.; Wiechmann, A.; Mussabekova, A.; Moody, S.; Simon, M.F.; Moulin, L.; Elliott, G.N.; Lacercat-Didier, L.; DaSilva, C.; et al. Endemic Mimosa species from Mexico prefer alphaproteobacterial rhizobial symbionts. New Phytol. 2016, 209, 319–333. [Google Scholar] [CrossRef]
- Elliott, G.N.; Chou, J.-H.; Chen, W.-M.; Bloemberg, G.V.; Bontemps, C.; Martínez-Romero, E.; Velázquez, E.; Young, J.P.W.; Sprent, J.I.; James, E. Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ. Microbiol. 2009, 11, 762–778. [Google Scholar] [CrossRef]
- Lardi, M.; De Campos, S.B.; Purtschert, G.; Eberl, L.; Pessi, G. Competition experiments for legume infection identify Burkholderia phymatum as a highly competitive β-Rhizobium. Front. Microbiol. 2017, 8, 1527. [Google Scholar] [CrossRef]
- Elliott, G.N.; Chen, W.-M.; Chou, J.-H.; Wang, H.-C.; Sheu, S.-Y.; Perin, L.; Reis, V.M.; Moulin, L.; Simon, M.F.; Bontemps, C.; et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 2007, 173, 168–180. [Google Scholar] [CrossRef] [PubMed]
- De Campos, S.B.; Lardi, M.; Gandolfi, A.; Eberl, L.; Pessi, G. Mutations in two Paraburkholderia phymatum Type VI secretion systems cause reduced fitness in interbacterial competition. Front. Microbiol. 2017, 8, 2473. [Google Scholar] [CrossRef] [PubMed]
- Morett, E.; Buck, M. In vivo studies on the interaction of RNA polymerase-σ54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters: The role of NifA in the formation of an open promoter complex. J. Mol. Biol. 1989, 210, 65–77. [Google Scholar] [CrossRef]
- Francke, C.; Kormelink, T.G.; Hagemeijer, Y.; Overmars, L.; Sluijter, V.; Moezelaar, R.; Siezen, R.J. Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genom. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Lardi, M.; Liu, Y.; Purtschert, G.; De Campos, S.B.; Pessi, G. Transcriptome analysis of Paraburkholderia phymatum under nitrogen starvation and during symbiosis with Phaseolus vulgaris. Genes 2017, 8, 389. [Google Scholar] [CrossRef]
- Lardi, M.; Liu, Y.; Giudice, G.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomics and transcriptomics identify multiple downstream targets of Paraburkholderia phymatum σ54 during symbiosis with Phaseolus vulgaris. Int. J. Mol. Sci. 2018, 19, 1049. [Google Scholar] [CrossRef]
- Sheu, S.-Y.; Chou, J.-H.; Bontemps, C.; Elliott, G.N.; Gross, E.; dos Reis Junior, F.B.; Melkonian, R.; Moulin, L.; James, E.K.; Sprent, J.I.; et al. Burkholderia diazotrophica sp. nov., isolated from root nodules of Mimosa spp. Int. J. Syst. Evol. Microbiol. 2013, 63, 435–441. [Google Scholar] [CrossRef]
- Clark, D.J.; Maaløe, O. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 1967, 23, 99–112. [Google Scholar] [CrossRef]
- Miller, W.G.; Leveau, J.H.; Lindow, S.E. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 2000, 13, 1243–1250. [Google Scholar] [CrossRef]
- Ong, C.-L.Y.; Beatson, S.A.; McEwan, A.G.; Schembri, M.A. Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl. Environ. Microbiol. 2009, 75, 6783–6791. [Google Scholar] [CrossRef]
- Pessi, G.; Ahrens, C.H.; Rehrauer, H.; Lindemann, A.; Hauser, F.; Fischer, H.-M.; Hennecke, H. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant Microbe Interact. 2007, 20, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Moulin, L.; Klonowska, A.; Caroline, B.; Booth, K.; Vriezen, J.A.; Melkonian, R.; James, E.; Young, J.P.W.; Béna, G.; Hauser, L.; et al. Complete Genome sequence of Burkholderia phymatum STM815T, a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand. Genom. Sci. 2014, 9, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.; Szklarczyk, D.; Trachana, K.; Roth, A.; Kuhn, M.; Muller, J.; Arnold, R.; Rattei, T.; Letunic, I.; Doerks, T.; et al. eggNOG v3.0: Orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012, 40, D284–D289. [Google Scholar] [CrossRef]
- Ledebur, H.C.; Gu, B.; Sojda, J.; Nixon, B.T. Rhizobium meliloti and Rhizobium leguminosarum dctD gene products bind to tandem sites in an activation sequence located upstream of sigma 54-dependent dctA promoters. J. Bacteriol. 1990, 172, 3888–3897. [Google Scholar] [CrossRef]
- Valentini, M.; Storelli, N.; Lapouge, K. Identification of C4-dicarboxylate transport systems in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2011, 193, 4307–4316. [Google Scholar] [CrossRef]
- Poole, P.; Allaway, D. Carbon and nitrogen metabolism in Rhizobium. Adv. Microb. Physiol. 2000, 43, 117–163. [Google Scholar] [CrossRef]
- Filloux, A. The rise of the Type VI secretion system. F1000Prime Rep. 2013, 5, 5. [Google Scholar] [CrossRef]
- Li, Y.G.; Hu, B.; Christie, P.J. Biological and structural diversity of type IV secretion systems. Microbiol. Spectr. 2019, 7, 1–15. [Google Scholar] [CrossRef]
- Liu, Y.; Bellich, B.; Hug, S.; Eberl, L.; Cescutti, P.; Pessi, G. The exopolysaccharide cepacian plays a role in the establishment of the Paraburkholderia phymatum—Phaseolus vulgaris symbiosis. Front. Microbiol. 2020, 11, 1600. [Google Scholar] [CrossRef]
- Garcia, E.; Bancroft, S.; Rhee, S.G.; Kustu, S. The product of a newly identified gene, gInF, is required for synthesis of glutamine synthetase in Salmonella. Proc. Natl. Acad. Sci. USA 1977, 74, 1662–1666. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wu, G.; Liao, Y.; Zeng, Q.; Wang, H.; Liu, F. RpoN1 and RpoN2 play different regulatory roles in virulence traits, flagellar biosynthesis, and basal metabolism in Xanthomonas campestris. Mol. Plant Pathol. 2020, 21, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Saldías, M.S.; Lamothe, J.; Wu, R.; Valvano, M.A. Burkholderia cenocepacia requires the RpoN sigma factor for biofilm formation and intracellular trafficking within macrophages. Infect. Immun. 2008, 76, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Kullik, I.; Fritsche, S.; Knobel, H.; Sanjuan, J.; Hennecke, H.; Fischer, H.M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the σ54 gene (rpoN). J. Bacteriol. 1991, 173, 1125–1138. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Mo, Z.-L.; Xiao, P.; Pan, H.-J.; Lan, X.; Li, G.-Y. Role of alternative sigma factor 54 (RpoN) from Vibrio anguillarum M3 in protease secretion, exopolysaccharide production, biofilm formation, and virulence. Appl. Microbiol. Biotechnol. 2012, 97, 2575–2585. [Google Scholar] [CrossRef] [PubMed]
- Hayrapetyan, H.; Tempelaars, M.; Groot, M.N.; Abee, T. Bacillus cereus ATCC 14579 RpoN (sigma 54) is a pleiotropic regulator of growth, carbohydrate metabolism, motility, biofilm formation and toxin production. PLoS ONE 2015, 10, e0134872. [Google Scholar] [CrossRef]
- Cai, Z.; Liu, Y.; Chen, Y.; Yam, J.K.H.; Chew, S.C.; Chua, S.L.; Wang, K.; Givskov, M.; Yang, L. RpoN regulates virulence factors of Pseudomonas aeruginosa via modulating the PqsR quorum sensing regulator. Int. J. Mol. Sci. 2015, 16, 28311–28319. [Google Scholar] [CrossRef]
- Lardi, M.; Aguilar, C.; Pedrioli, A.; Omasits, U.; Suppiger, A.; Cárcamo-Oyarce, G.; Schmid, N.; Ahrens, C.H.; Eberl, L.; Pessi, G. σ54-dependent response to nitrogen limitation and virulence in Burkholderia cenocepacia strain H111. Appl. Environ. Microbiol. 2015, 81, 4077–4089. [Google Scholar] [CrossRef]
- Salazar, E.; Díaz-Mejía, J.J.; Moreno-Hagelsieb, G.; Martínez-Batallar, G.; Mora, Y.; Mora, J.; Encarnación, S. Characterization of the NifA-RpoN regulon in Rhizobium etli in free life and in symbiosis with Phaseolus vulgaris. Appl. Environ. Microbiol. 2010, 76, 4510–4520. [Google Scholar] [CrossRef]
- Hauser, F.; Pessi, G.; Friberg, M.; Weber, C.; Rusca, N.; Lindemann, A.; Fischer, H.-M.; Hennecke, H. Dissection of the Bradyrhizobium japonicum NifA+σ54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol. Genet. Genomics 2007, 278, 255–271. [Google Scholar] [CrossRef]
- Fazli, M.; Rybtke, M.; Steiner, E.; Weidel, E.; Berthelsen, J.; Groizeleau, J.; Bin, W.; Zhi, B.Z.; Yaming, Z.; Kaever, V.; et al. Regulation of Burkholderia cenocepacia biofilm formation by RpoN and the c-di-GMP effector BerB. Microbiologyopen 2017, 6, e00480. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.J.; Golby, P.; Omrani, D.; Broad, S.A.; Harrington, V.L.; Guest, J.R.; Kelly, D.J.; Andrews, S.C. Inactivation and regulation of the aerobic C4-dicarboxylate transport (dctA) gene of Escherichia coli. J. Bacteriol. 1999, 181, 5624–5635. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.J.; Poole, P.S. Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J. Bacteriol. 1998, 180, 2660–2669. [Google Scholar] [CrossRef] [PubMed]
- Finan, T.M.; Wood, J.M.; Jordan, D.C. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J. Bacteriol. 1983, 154, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Ronson, C.W.; Astwood, P.M.; Downie, J.A. Molecular cloning and genetic organization of C4-dicarboxylate transport genes from Rhizobium leguminosarum. J. Bacteriol. 1984, 160, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Engelke, T.; Jagadish, M.N.; Pühler, A. Biochemical and genetical analysis of Rhizobium meliloti mutants defective in C4-dicarboxylate transport. Microbiology 1987, 133, 3019–3029. [Google Scholar] [CrossRef][Green Version]
- Müller, J.; Boller, T.; Wiemken, A. Trehalose becomes the most abundant non-structural carbohydrate during senescence of soybean nodules. J. Exp. Bot. 2001, 52, 943–947. [Google Scholar] [CrossRef]
- Streeter, J.G. Effect of trehalose on survival of Bradyrhizobium japonicum during desiccation. J. Appl. Microbiol. 2003, 95, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Streeter, J.G.; López-Gómez, M.L. Three enzymes for trehalose synthesis in Bradyrhizobium cultured bacteria and in bacteroids from soybean nodules. Appl. Environ. Microbiol. 2006, 72, 4250–4255. [Google Scholar] [CrossRef]
- Cytryn, E.J.; Sangurdekar, D.P.; Streeter, J.G.; Franck, W.L.; Chang, W.-S.; Stacey, G.; Emerich, D.W.; Joshi, T.; Xu, D.; Sadowsky, M.J. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J. Bacteriol. 2007, 189, 6751–6762. [Google Scholar] [CrossRef]
- Sugawara, M.; Cytryn, E.J.; Sadowsky, M.J. Functional role of Bradyrhizobium japonicum trehalose biosynthesis and metabolism genes during physiological stress and nodulation. Appl. Environ. Microbiol. 2010, 76, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Lardi, M.; Murset, V.; Fischer, H.-M.; Mesa, S.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomic profiling of Bradyrhizobium diazoefficiens-induced root nodules reveals both host plant-specific and developmental signatures. Int. J. Mol. Sci. 2016, 17, 815. [Google Scholar] [CrossRef] [PubMed]
- Suárez, R.; Wong, A.; Ramirez, M.; Barraza, A.; Orozco, M.D.C.; Cevallos, M.A.; Lara, M.; Hernández, G.; Iturriaga, G. Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in Rhizobia. Mol. Plant Microbe Interact. 2008, 21, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef]
- Delorge, I.; Figueroa, C.M.; Feil, R.; Lunn, J.E.; Van Dijck, P. Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem. J. 2015, 466, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Gonzalez-Uriarte, A.; Griffiths, C.A.; Hassani-Pak, K. The Role of trehalose 6-phosphate in crop yield and resilience. Plant Physiol. 2018, 177, 12–23. [Google Scholar] [CrossRef]
- Hengge-Aronis, R.; Klein, W.; Lange, R.; Rimmele, M.; Boos, W. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 1991, 173, 7918–7924. [Google Scholar] [CrossRef]
- Flechard, M.; Fontenelle, C.; Blanco, C.; Goude, R.; Ermel, G.; Trautwetter, A. RpoE2 of Sinorhizobium meliloti is necessary for trehalose synthesis and growth in hyperosmotic media. Microbiology 2010, 156, 1708–1718. [Google Scholar] [CrossRef]
- Bernard, C.S.; Brunet, Y.R.; Gavioli, M.; Lloubès, R.; Cascales, E. Regulation of type VI secretion gene clusters by σ54 and cognate enhancer binding proteins. J. Bacteriol. 2011, 193, 2158–2167. [Google Scholar] [CrossRef]
- Sana, T.G.; Soscia, C.; Tonglet, C.M.; Garvis, S.; Bleves, S. Divergent control of two type VI secretion Systems by RpoN in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e76030. [Google Scholar] [CrossRef]
- Sheng, L.; Gu, D.; Wang, Q.; Liu, Q.; Zhang, Y. Quorum sensing and alternative sigma factor RpoN regulate type VI secretion system I (T6SSVA1) in fish pathogen Vibrio alginolyticus. Arch. Microbiol. 2012, 194, 379–390. [Google Scholar] [CrossRef]
- Dong, T.G.; Mekalanos, J.J. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012, 40, 7766–7775. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).