1. Introduction
Skin cancer is the most common cancer, with one in five people suffering from it once in their life [
1]. Skin cancers are broadly classified as melanoma and non-melanoma skin cancers, which are further classified as basal cell carcinoma and squamous cell carcinoma [
1]. Once the lesion moves toward the lower layers of the skin it is almost impossible to prevent metastasis, which makes skin cancer the deadliest form of cancer [
2]. Hence, to improve the rate of survival of the patients, it is crucial to identify these malignancies as early as possible, and provide the necessary treatment to prevent them from spreading and recurring [
3]. Most of the diagnoses of skin cancer are based on the ABCDE rule, where the physical aspects of the area are analyzed by considering asymmetry, border, color, diameter, and evolution [
4].
There have been advancements in deep learning for skin cancer detection and classification, but the generalization across different image conditions, skin tones, and noise in the data remains one of the major challenges. Also, it is essential to ensure that the boundary is detected accurately during the segmentation for it to have a positive impact on the model’s accuracy.
Ibrahim et al. [
5] and Anand et al. [
6] explored the use of transfer learning using the VGG16 backbone. Reis et al. [
7] proposed an InSiNet model which is based on the Inception module of GoogleNet. Gomathi et al. [
3] studied the applications of dual-optimization-based deep learning networks. Arabahmadi, Farahbakhsh, and Rezazadeh [
8] discussed the use of semantic segmentation to identify the parts of the image, the use of architectures like U-Net, and convolutional neural networks for medical image classification and discussed the advantages and disadvantages of different pre-trained architectures like GoogleNet, VGGNet, ResNet, etc. These studies show that significant progress has been made in developing deep learning models to detect and classify skin cancer, but there is still room for improvement in the model’s performance on previously unseen data.
In our study, we explored the use of the ISIC 2016 dataset [
9] to train the segmentation model. We used the model to segment and outline the lesion region in the Kaggle dataset of 10,000 skin lesion images that have been collected from different ISIC directories [
10]. We explored the use of different pre-processing techniques, like augmentation, normalization, and U-Net segmentation, to enhance the model’s robustness and accuracy. We have developed a classification model that uses transfer learning, using ResNet-50 as a backbone. The performance of the model was tested using different metrics. It achieved an accuracy of 0.9280, precision of 0.9864, and recall of 0.8680. The major contributions of this paper are as follows:
We applied the canny edge detection technique to delineate the lesion area from the segmented image, which allowed us to visualize the features of the region of interest in the image.
We integrated the results of the segmentation model to highlight the lesion area in the image that is later passed to the classification model. This improved the ability of the classifier to detect and classify the lesion as either benign or malignant.
We used transfer learning, with ResNet-50 as the backbone for the classification model, demonstrating that we can adopt a pre-trained model for skin cancer classification, while achieving a high accuracy.
We have divided the study into the following sections:
Section 2 contains the literature review.
Section 3 contains the theoretical background.
Section 4 discusses materials and methods.
Section 5 describes the experimental design and results.
Section 6 discusses the results of the study, which is concluded in
Section 7.
2. Literature Review
Ibrahim et al. [
5] proposed a transfer learning model using VGG16 for skin cancer classification and obtained an accuracy of 84.242%. They highlighted that transfer learning from a deeper network reduces the use of computational resources needed to achieve a model with a high accuracy. They implemented data augmentations to improve robustness, extracted feature maps from different layers of VGG16, and passed them through fully connected layers to obtain higher accuracy.
Anand et al. [
6] proposed a transfer learning model using the VGG16 pre-trained model. They used different augmentation techniques, like rotation, flipping, and brightening. They finetuned the VGG16 model by adding a flattened layer and a dense layer, and used Leaky ReLU to improve the accuracy of the model. They have compared the performance of the VGG16 and finetuned it using different batch sizes. They achieved good classification results, but there was still the possibility for other pre-processing and augmentation techniques to be used, and for testing with other pre-trained networks to improve the performance of the classification models.
Sivakumar et al. [
11] proposed an automatic deep learning model that uses a Convolutional Neural Network architecture with ResNet50 to identify skin cancer as either malignant or benign. In their experiments, they tested using different models like ResNet50, CNN, CV-CNN, and SqueezeNet. They have underscored that their proposed model has an improved classification accuracy because of the noise reduction and enhancement capability of spectral image information of the model. However, we could look further into handling noisy data and testing models across different skin tones for generalizing models across different populations.
Abdelhafeez et al. [
12] implemented Neutrosophic c-means clustering (NCMC) and Fuzzy c-means Clustering (FCMC) to detect skin cancer and found that NCMC performed better. They aimed to resolve the shortcomings of the traditional models due to lighting and chrominance. They found that NCMC managed the unclear data points better and improved the performance of clustering. This approach performed better than the mean shift and K-means clustering approaches.
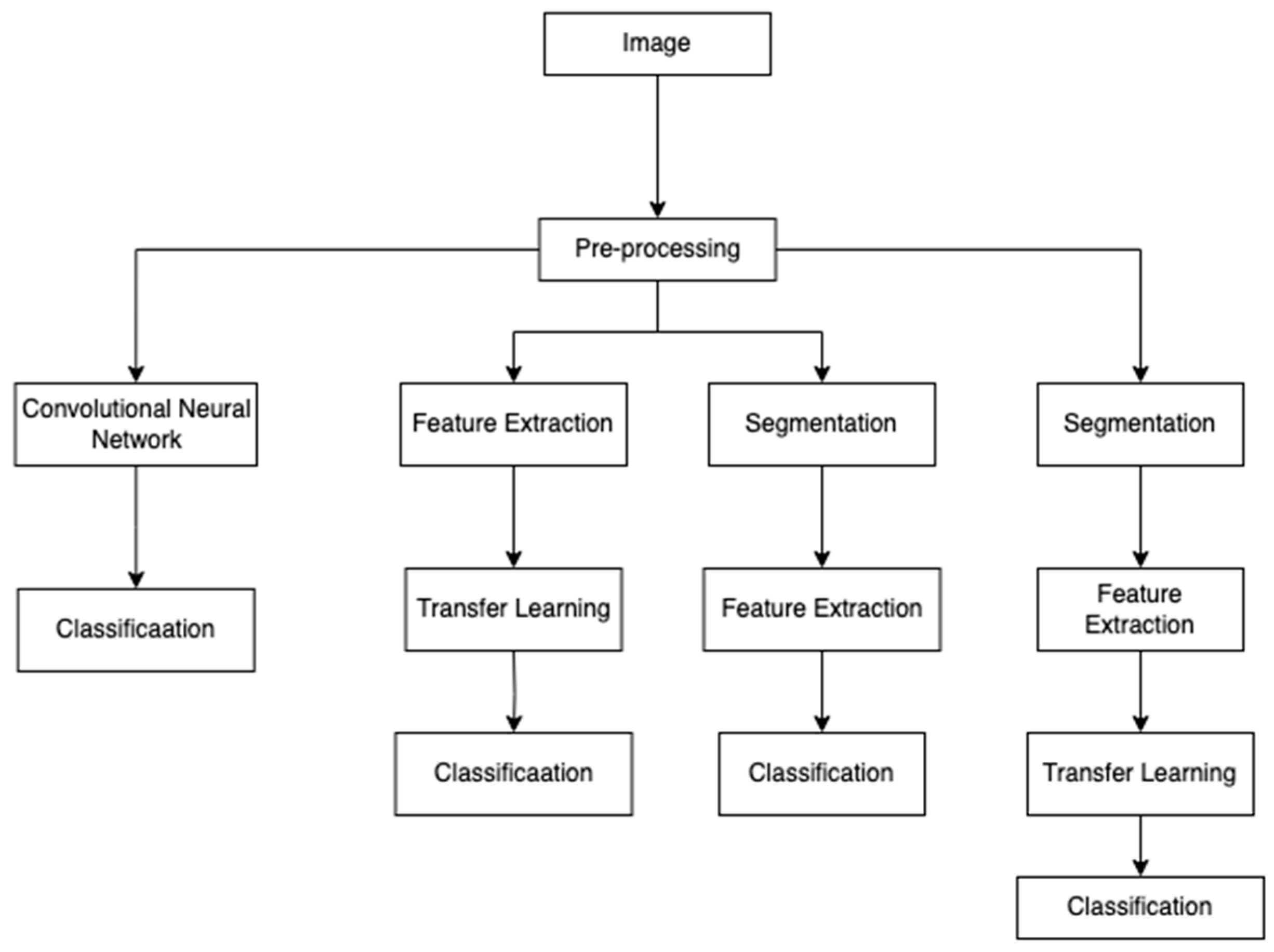
Figure 1 shows the approaches taken by the different studies using deep learning and machine learning techniques for skin cancer classification problems. The images are pre-processed using methods like resizing, augmentation, normalization, etc. ([
5,
11]), to ensure that the models generalize well in images of varying conditions, skin tones, etc. After this, the next steps taken are feature extraction or segmentation. Ibrahim et al. [
5] and Anand et al. [
6] implemented the VGG16 model to extract the features before the classification. Sivakumar et al. [
11] implemented segmentation to identify and focus on the lesion area from the surrounding skin. After this, the pre-trained models like VGG16, ResNet, etc., were finetuned to improve the classification accuracy.
We implemented different image pre-processing steps on each image. We used this approach to ensure that each batch has a different variation in the same image to make the model robust to variations in lighting, shape, and texture. We have integrated U-Net-based segmentation to isolate lesion regions and use it to delineate the lesion area. We did this to reduce the influence of background noise and enhance feature extraction to improve classification accuracy. We used pre-trained ResNet50 as our base model. We updated ResNet’s fully connected layer with the new layers tailored to our case.
Existing methods use segmentation and classification as separate tasks [
13] or focus on pre-trained networks [
6] that learn representations based on the entire image. We integrated segmentation results into our classification pipeline. We separated the lesion area using the segmented image as a guide and a canny edge detection method to delineate the region. We used the images with delineated lesion areas as input to the classification model to improve the model’s accuracy by focusing on the lesion area.
Our work integrated segmentation in pre-processing and modified the network architecture of ResNet50 to improve the accuracy of the skin cancer classification, which differentiates our work from other’s works.
3. Theoretical Background
In this section, we discuss the different concepts that have been used in this study.
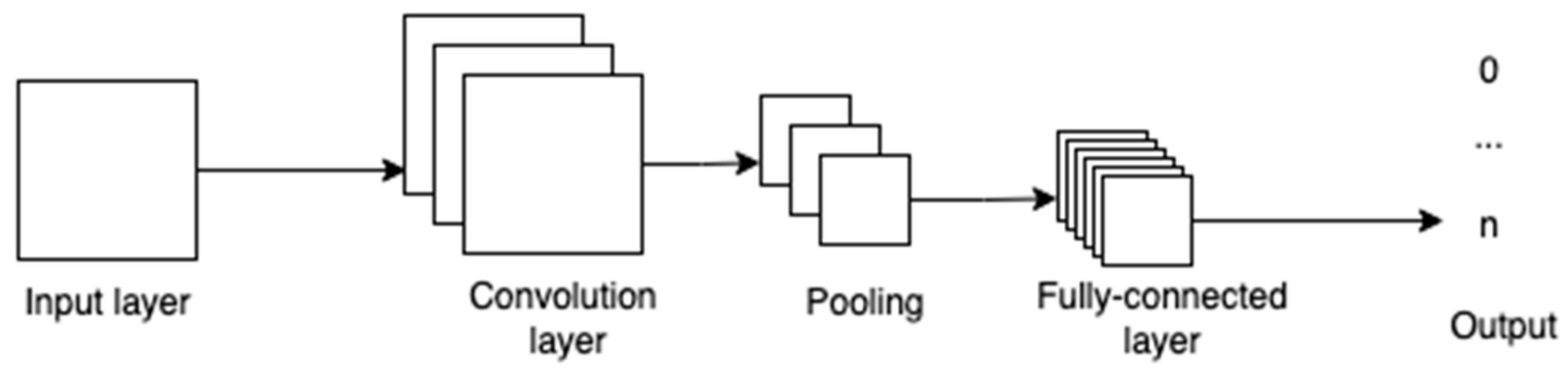
3.1. Convolutional Neural Network
CNNs are networks whose structure is guided by the biological nervous system with multiple nodes that are connected to each other [
14]. Each node receives an input and carries out calculations to obtain an output. Between the input and the output layer, there can be one or more hidden layers. These layers act as the decision-making layers, as they are responsible for comparing how a change improves or worsens the final output.
Figure 2 shows the various layers in a convolutional neural network. The convolution layer takes in the pixels and its outputs are fed to the pooling layer, where specific parts of the image, like edges, corners, etc., are identified, and are then fed to the fully connected layers to obtain the final output.
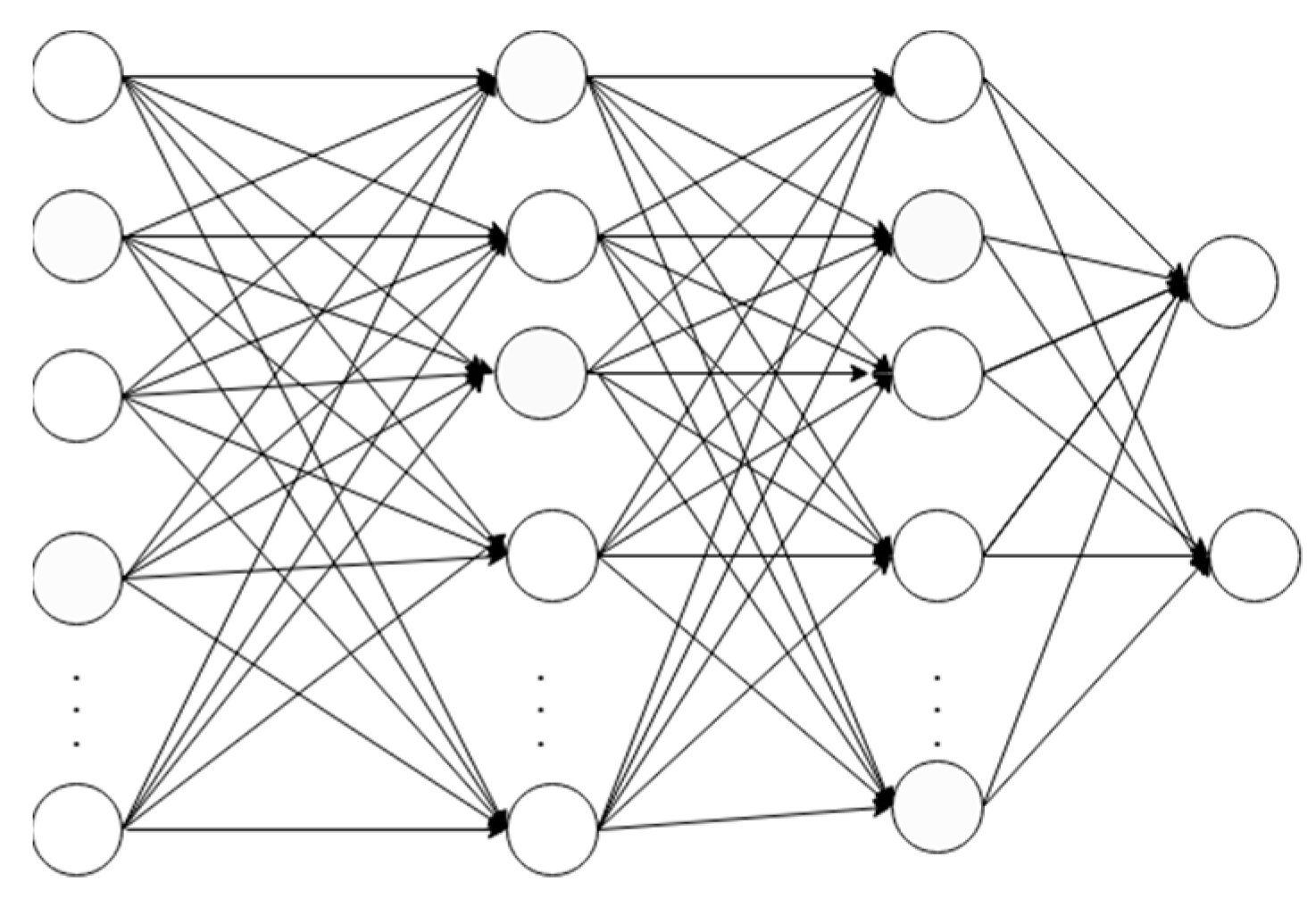
3.2. Feedforward Network
In a feedforward network, the vector of multiple dimensions is fed as an input which then passes through a number of hidden layers. While traversing through the hidden layers, the decisions from the previous layers are made. The decision is based on the calculations on how the stochastic updates by themselves affect the accuracy of the output. This process is called learning [
15].
Figure 3 shows a simple four-layered feedforward neural network. It has an input layer, two hidden layers, and an output layer with two classes.
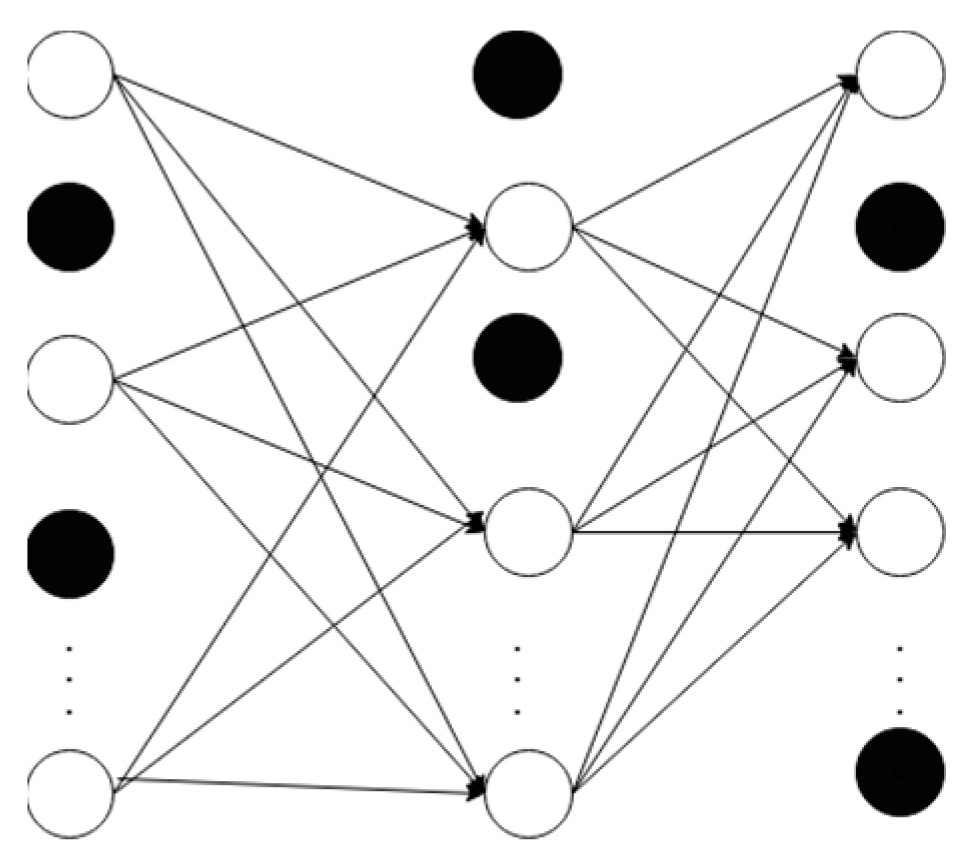
3.3. Dropout
Dropout is a technique where we randomly shut down some neurons, like 40% of the neurons in
Figure 4. This step reduces the probability of overfitting and improves generalization [An Introduction to Neural Networks and Deep Learning, 2017].
Figure 4 shows a neural network with a dropout rate of 40%, where the deactivated nodes are represented by the darkened nodes.
3.4. U-Net
Ronneberger, Fischer, and Brox [
16] proposed a U-Net architecture for segmentation. The segmentation is performed in two phases, with down-sampling followed by up-sampling. In the down-sampling phase of the architecture, the spatial resolution of the image is reduced and the number of feature channels is increased. Each step in this phase has a convolution layer, followed by a ReLU activation; max pooling with a 2 × 2 filter is used to reduce the spatial dimension by half.
The up-sampling phase reverses the down-sampling phase, where the spatial resolution of the image is restored while preserving the learned features. It has a 2 × 2 up-convolution, followed by two 3 × 3 convolution and a ReLU activation, to refine the output. In the last layer, there is a 1 × 1 convolution, which is the segmentation mask, to map the feature maps to the desired number of output classes, which are the class labels.
3.5. Canny Edge Detection
A canny edge filter is a filter that detects edges in multiple stages, which is based on first derivative of a Gaussian function. This performance of the algorithm is based on its higher probability of marking the true edge points, the points of the predicted edges being as close to the true edge’s center as possible, and on the fact that it provides one edge point for one edge [
17]. In this algorithm, the Gaussian filter is used at first to reduce the noise, then the intensity gradient is calculated to identify the direction and strength of edges. After this, the double threshold is applied to identify the potential edges, where the edges are identified as strong (gradient value > high threshold) and weak (low threshold < gradient value < high threshold). This is followed by the tracking of edges, where the strong edges and the weak edges connected to them are considered edges.
3.6. ResNet-50
ResNet50 is a residual network architecture that implements residual learning using skip connections. It is a type of ResNet which has a depth of 50 layers. It has 49 convolution layers, followed by a fully connected layer. It has a set of residual blocks where the convolution operations are carried out to transform the input and the output is added back to the input to form the residual connection. Each of these blocks uses a skip connection to skip certain layers for identity mapping, which allows the block to learn residual functions instead of full mapping. This simplifies the optimization of deep networks [
18].
3.7. Transfer Learning
Transfer learning is an ML technique, where a model developed for a specific task is used as a starting point for a related task [
19].
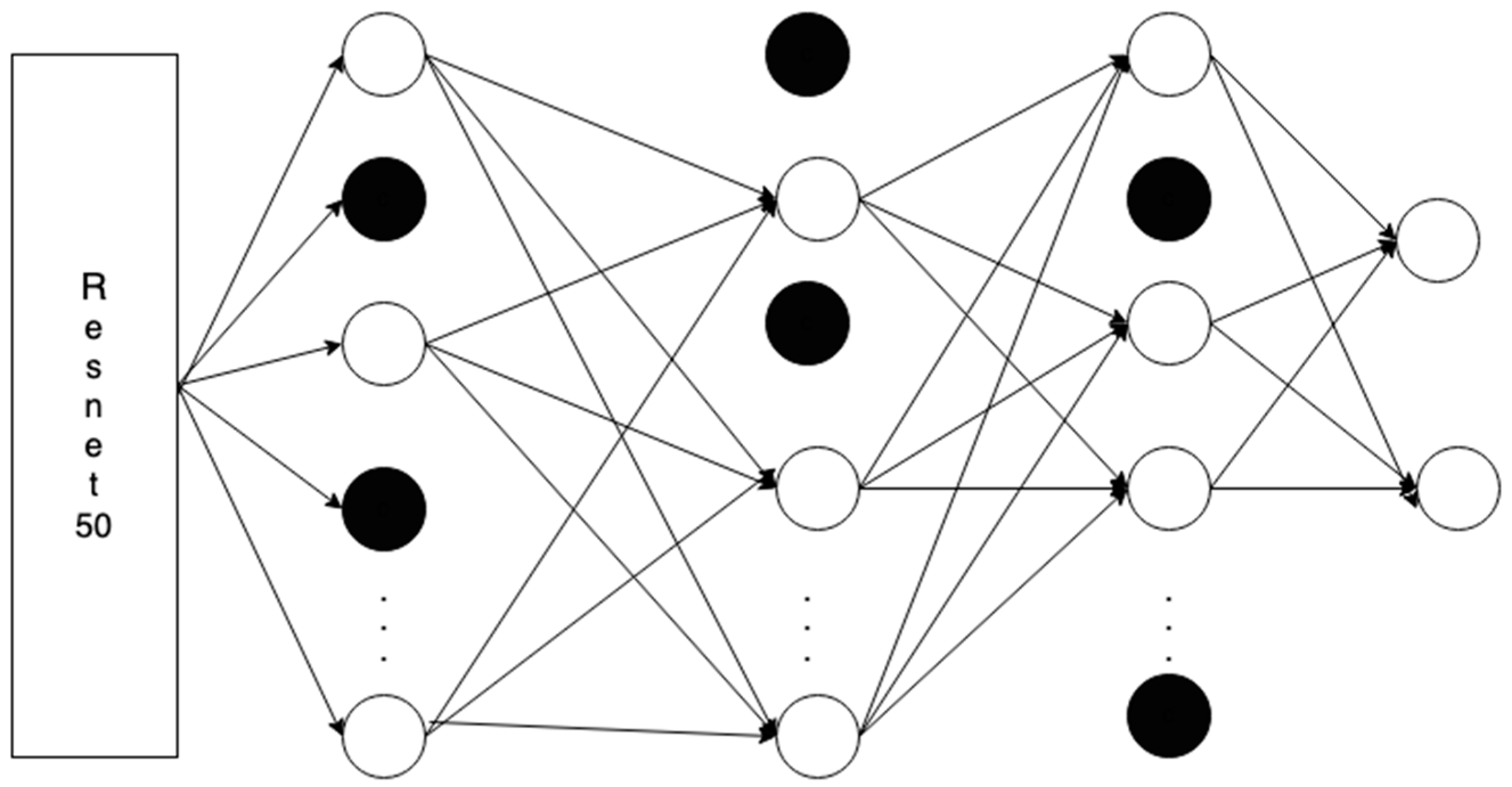
Figure 5 illustrates the architecture used in our approach, which combines a ResNet50 backbone with a feedforward neural network. The benefit of transfer learning is in the initial performance of the model, improving the learning speed of the model, and also improving the performance of the model, as it starts from a more advanced point.
4. Materials and Methods
In this section we discuss the data we have used in our study, the steps we implemented, and visualize the results of those steps.
4.1. Dataset
We have used the ISIC 2016 dataset [
9] to train the segmentation model. We have used 900 skin lesion images and their respective masks to train the U-Net segmentation model. We have used 379 skin lesion images and their masks to test our segmentation model. The original lesion images are in JPG format and the ground truth masks are in PNG format, all in a resolution of 1022 × 767 pixels. The ground truth images are black and white, colored where the white portion indicates the lesion portion in the image.
We have used the melanoma skin cancer dataset of 10,000 images from Kaggle [
10] to train our classification model. The dataset has 5000 benign and 4605 malignant images for training. It has 1000 test images with 500 benign and malignant each, respectively. The images are in a JPG format, with a resolution of 300 × 300 pixels.
4.2. Data Preprocessing
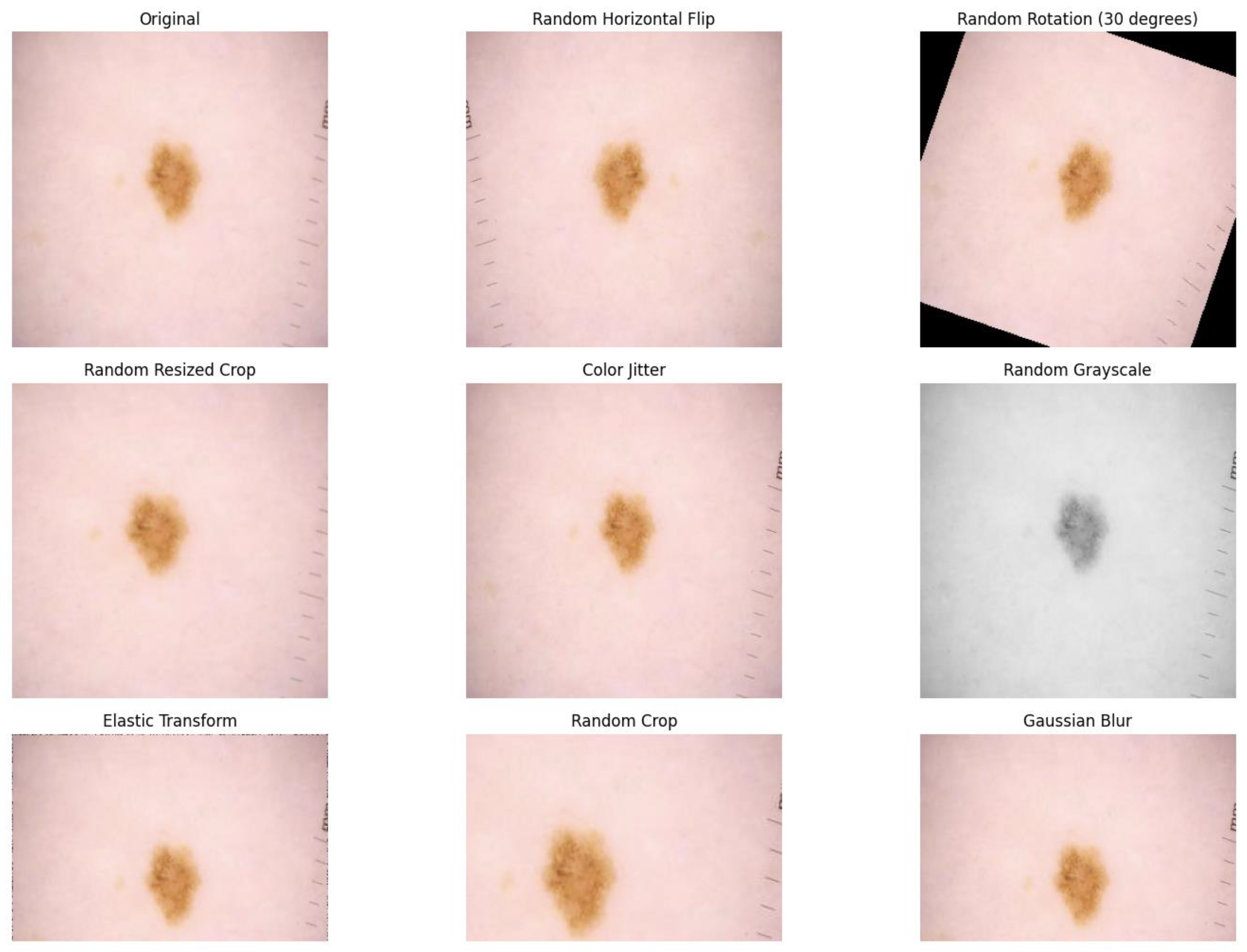
We resized the images to 224 × 224 pixels and employed different pre-processing and augmentation techniques to improve the performance of the model in unseen data. We have implemented random horizontal flipping, random rotation of 30 degrees, random resized crop with the crop factor of 0.8 in the height, color-jitter to simulate different lighting conditions, random gray-scaling, random cropping, and random Gaussian blur. The sample of changes in the lesion image after the pre-processing steps is shown in
Figure 6.
To standardize the input images, we normalized them. We have used the values [0.485, 0.456, 0.406] which is the mean of the pixel values, and [0.229, 0.224, 0.225] which is the standard deviation of the pixel values for red, green, and blue color channels, as these are the mean and standard deviation of pixel values for ImageNet dataset.
We have applied the transformations on-the-fly during the training. Because of this approach, the number of images after pre-processing is the same as in the training dataset, i.e., 10,000. However, each batch will see different variations in the same image.
4.3. Segmentation
We paired the lesion images and their corresponding segmentation mask and applied the same augmentation and pre-processing steps to both the lesion image and the segmentation mask. We have resized the images to 224 × 224 pixels and used pre-processing steps which consist of flipping, rotation, resized cropping, and gray-scaling. The training of the segmentation model was performed in a batch size of 4 for 30 epochs.
In our study, it was very important to delineate the region of interest so that the classifier focuses more on the relevant region to improve the classification accuracy. So, we used the U-Net segmentation method, as it has been used in numerous medical image segmentation tasks, where the separation of the region of interest is important. This segmentation model in our application is structured in an encoder and decoder phase. In the encoder phase, we used a block with two convolutional layers, each followed by a ReLU activation. Each layer used a kernel size of 3 × 3, with padding of 1. Five down-sampling blocks were used. The number of feature channels doubled in each block, with 64 output features in the first block, to 1024 output features in the fifth block, as we went deeper. In the decoder phase, the deconvolution layers were used to up-sample the feature maps. In each deconvolution layer, the number of feature channels was reduced in each block, with 512 output channels in the first block to 64 output channels in the fourth block. The final output layer was a 1 × 1 convolution layer that reduced 64 output channels to 1 channel for binary segmentation, which represented the predicted mask for each pixel in the input image.
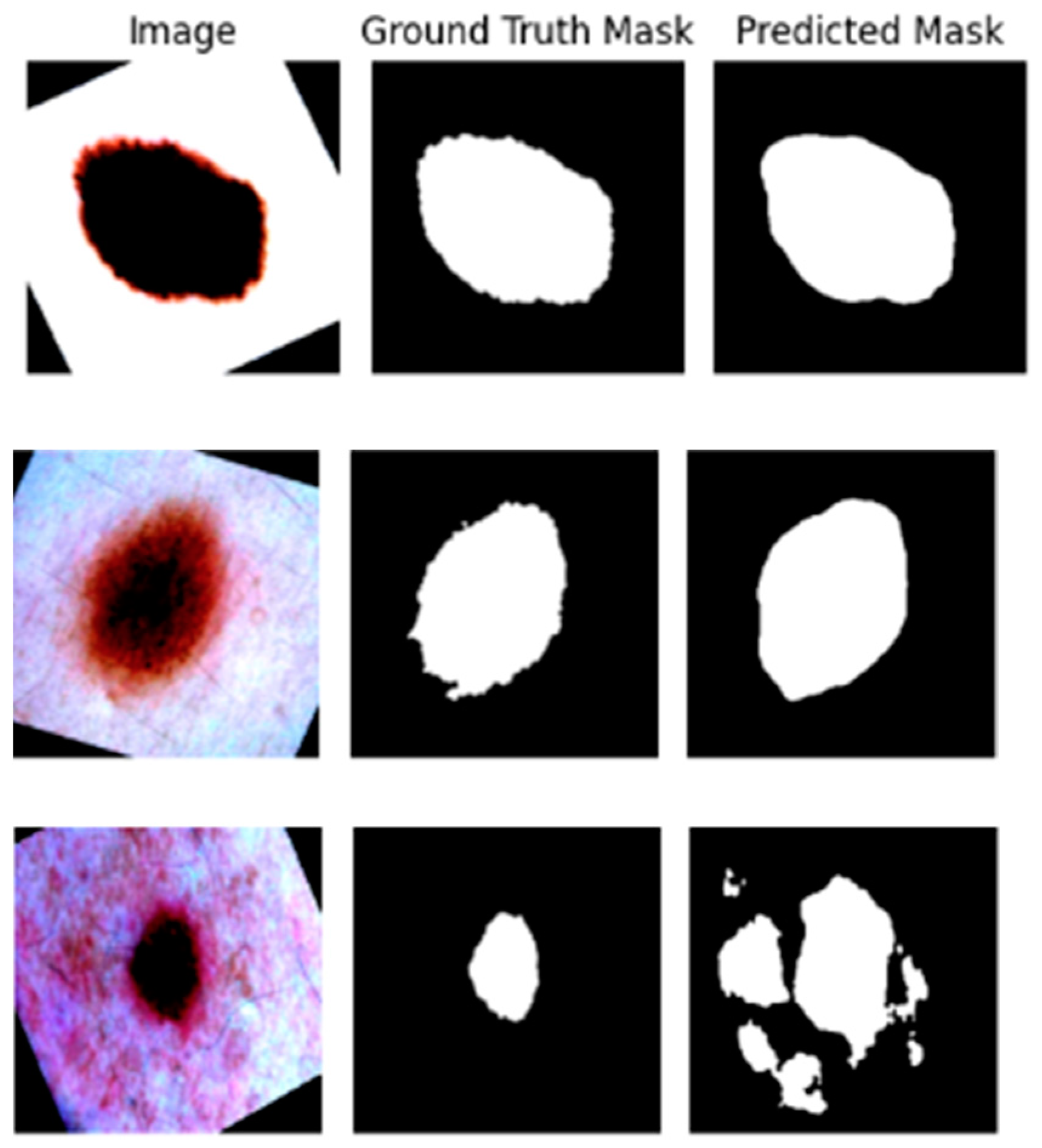
Figure 7 shows the original images in the first column, the ground truth images in the second column, and the images that are predicted by the segmentation model in the third column, which allowed us to visualize the results of the segmentation model.
4.4. Transfer Learning
We used the ResNet-50 pre-trained model as a backbone for transfer learning in our skin cancer classification dataset. We used the pre-trained model as a feature extractor for our classification dataset. We replaced the final fully connected layer in the ResNet-50 model with a customized layer for our binary classification task.
We implemented a multi-layer fully connected architecture. We used five layers of a feedforward neural network, which took the output features of ResNet-50 as input. The final output layer had two classes. For each of the fully connected layers, we used the ReLU activation function followed by the dropout layer. The dropout layer with a probability of 0.4 was added after the ReLU activation, which deactivated some neurons during training to reduce overfitting.
Initially, we froze the pre-trained layers to stop the weights of the pre-trained model from being updated during training. Once all the feedforward networks were added, we unfroze the layers to finetune the pre-trained model to the characteristics of skin cancer images. We trained this model using cross-entropy loss as the loss function and Adam optimizer to optimize the model parameters.
5. Experiment Design and Results
In this section we discuss the steps we took in conducting this study.
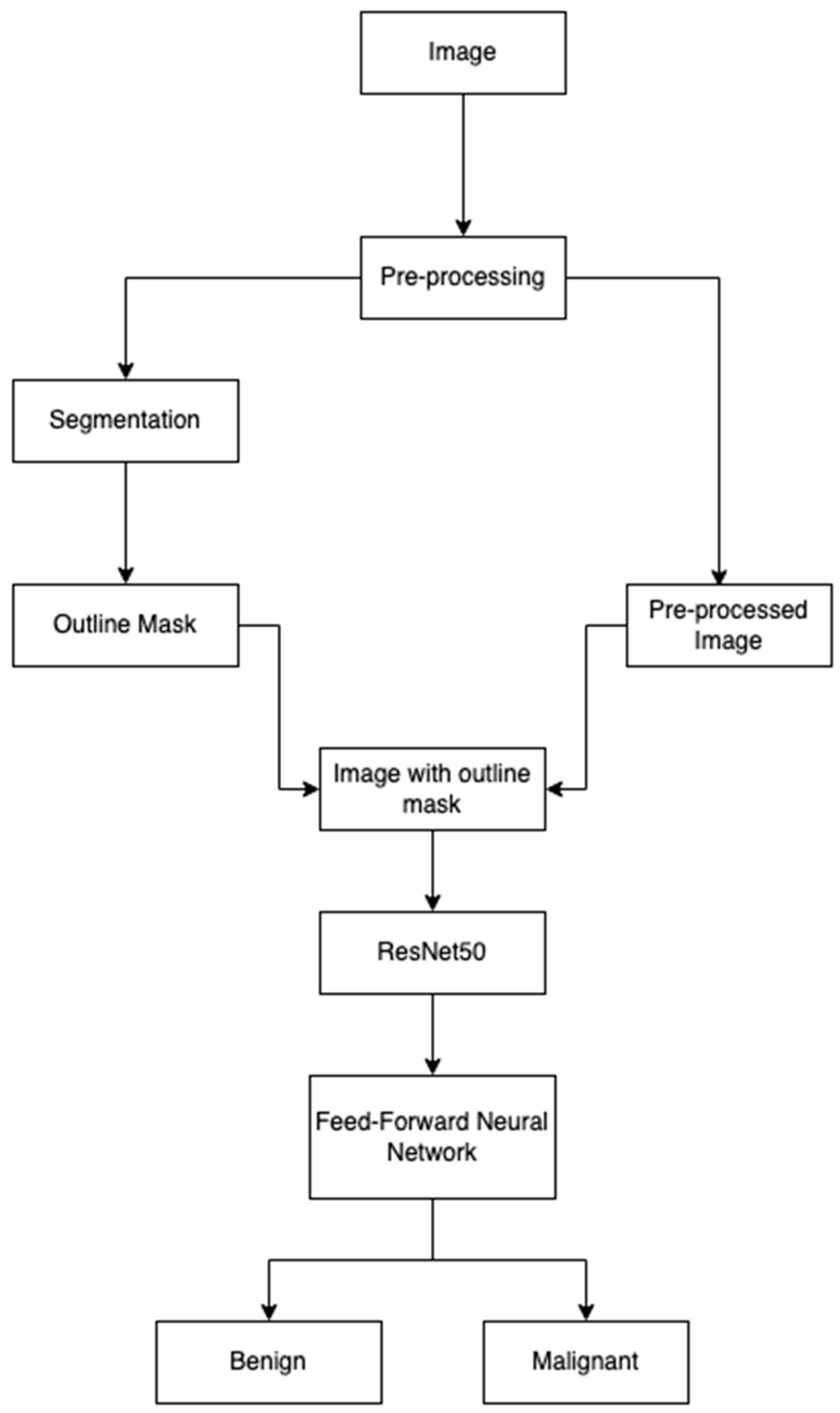
Figure 8 shows the steps we implemented. We used the 900 images from ISIC 2016 [
9] for training the segmentation model. We used 9605 images from the Kaggle dataset [
10] to train our classification model; 80% of the data were used for training and 20% for the validation during the training process. A total of 1000 test images from the dataset were used for testing the model. The segmentation model was trained for 30 epochs with a batch size of 4 and the classification model was trained for 20 epochs with the batch size of 32.
The training images for the classification model were first pre-processed and passed through the segmentation model. We applied the following pre-processing steps:
Resizing: All the images were resized to a fixed dimension of 224 × 224 pixels.
Random Horizontal Flip: The images were flipped at a probability of 0.5 to simulate different orientations of the lesions.
Random Rotation: Random rotations of up to 30 degrees were applied to the images to desensitize the model to the orientations of lesion areas in images.
Random Resized Crop: The images were cropped randomly and resized to 224 × 224 pixels to introduce variability to help the model learn more generalized features.
Color Jitter: This technique was used randomly to change the brightness, contrast, saturation, and hue of the image. This was carried out with the aim of making the model less sensitive to lighting conditions.
Random Grayscale: We used this technique to convert the image to grayscale with a probability of 0.1 to simulate variation in lighting conditions and imaging devices.
Elastic transform: This step was carried out to achieve random distortions in the image. This aimed to make the model more insensitive to small deformations that can happen in skin lesions.
Gaussian Blur: It was applied at random to simulate an out-of-focus image.
After these steps, the images were converted to PyTorch tensor format and normalized, so that the image pixel values were within the range suitable for model training.
All our images went through the pre-processing steps on-the-fly, and we used segmentation on all the images as an enhancement in the pre-processing step. Our workflow was designed to integrate segmentation as an enhancement to pre-processing, rather than as a distinct phase. The segmentation process serves as an intermediate step that refines the input data before classification. Thus, the obtained images were then fed to the classification model for training, which constituted of ResNet50 and a feedforward network, which classified the images as benign or malignant. We chose the ResNet-50 architecture because of its extensive use in the similar studies and the balance it provides between computational efficiency and accuracy. It demands fewer computational resources while achieving a very high accuracy.
We have also tuned hyperparameters like learning rate, optimizer selection, and dropout ratio to improve the model performance. We incorporated the Adam optimizer because it has shown good results in classification tasks in the previous studies. We started with the dropout ratio of 0.5 and reduced it by 0.1. We found that a dropout ratio of 0.4 was the best for our model accuracy. We also employed a learning scheduler to adjust the learning rate dynamically. We started with a learning rate of 0.0001. We scheduled it to reduce by a factor of 0.1 every 10 epochs to achieve rapid converges in the early epochs and to prevent overshooting optimal solutions.
The research work was carried out using the PyTorch 2.6.0 framework and the segmentation and classification models were trained in Google-Colab using the A100 GPU.
5.1. Metrics
The following metrics are used to evaluate the model. The terms used in the metrics are as follows:
True Positive (): Positive cases that are correctly predicted.
True Negative (): Negative cases that are correctly predicted.
False Positive (): Positive cases that are incorrectly predicted.
False Negative (): Negative cases that are incorrectly predicted.
5.1.1. Accuracy
It is the measure of how often the model’s predictions are correct.
5.1.2. Precision
It is the measure of how many of the positively classified cases are actually positive.
5.1.3. Recall
It is the measure of how many positive cases of all the positive cases were predicted correctly.
5.1.4. F1-Score
It is the harmonic mean of precision and recall, and is an important measure when precision and recall are important, like in our case.
5.1.5. Confusion Matrix
It is a 2 × 2 matrix that shows how much data from each class was classified into which category. It shows the number of TP, FP, TN, and FN. It allows us to visualize how a model performs in more detail.
6. Results
In this study, we have used different visual and quantitative results to validate the effectiveness of the skin cancer classification method we propose. The findings present the integration of the segmentation and classification tasks and the comparison of the obtained results with the results of the previous studies.
Figure 9 shows the pre-processed original dermoscopic images and the segmentation mask predicted by the U-Net segmentation model trained in the skin cancer dataset. The white portion in the segmented images represents the suspected lesions with a boundary between the skin and the lesion. The accuracy of the boundaries represents the ability of the segmentation model to separate the lesions and skin, which is expected to improve the accuracy of the classification task. We trained our segmentation model using the ISIC-2016 dataset [
9] and obtained an F1 score of 0.8965 and an Intersection over Union score of 0.7966. We achieved a test accuracy of 89.64%. This result suggests that our segmentation is capable of differentiating the boundaries of the lesion with minimal misclassification.
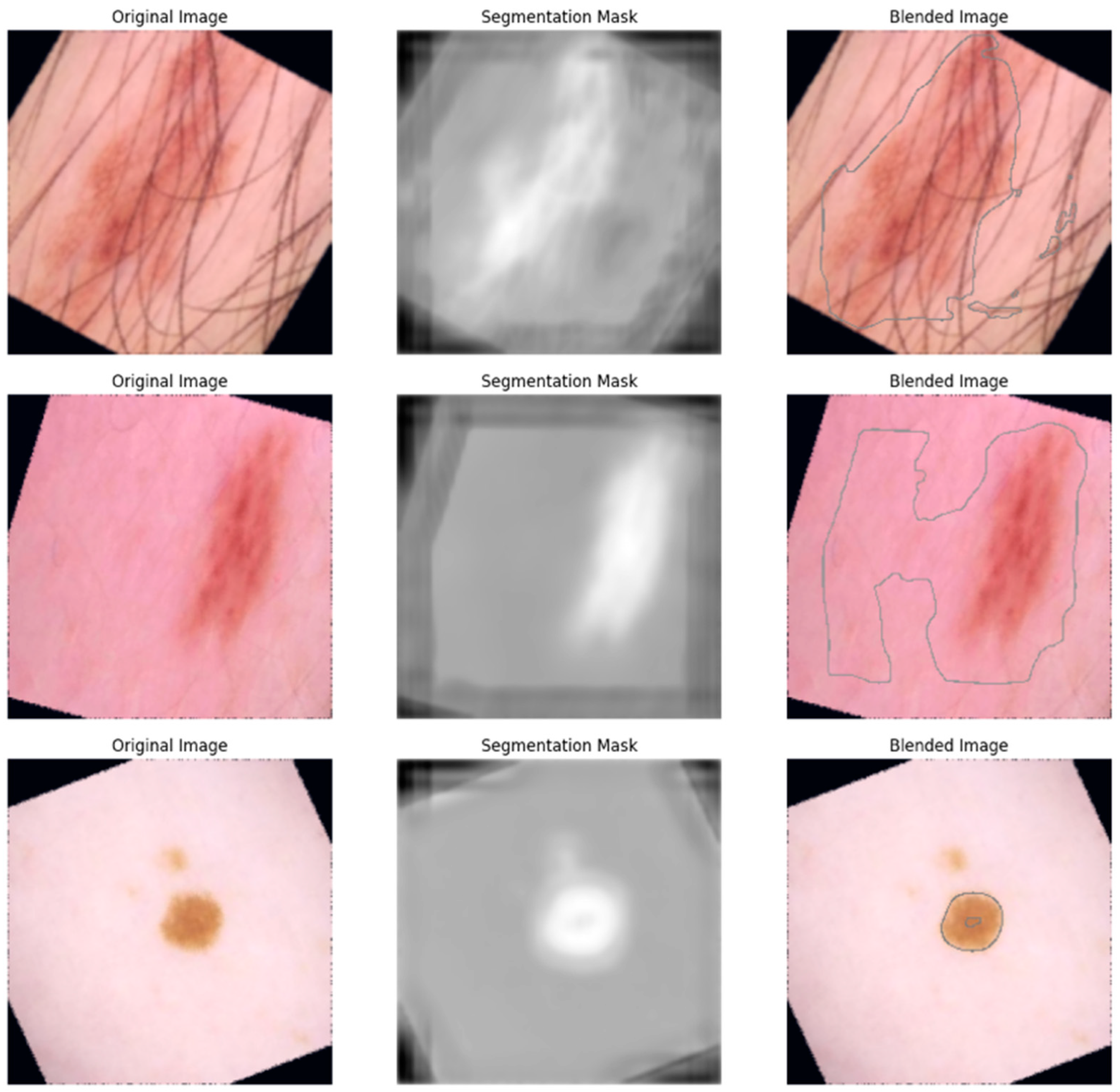
Figure 10 shows the blended images with an outlined lesion area which are obtained by overlaying the original images with their corresponding predicted segmentation mask outline. The visualization shows what the original images look like, their corresponding segmentation mask, and the final output of the blending, which shows the spatial localization capacity of the model.
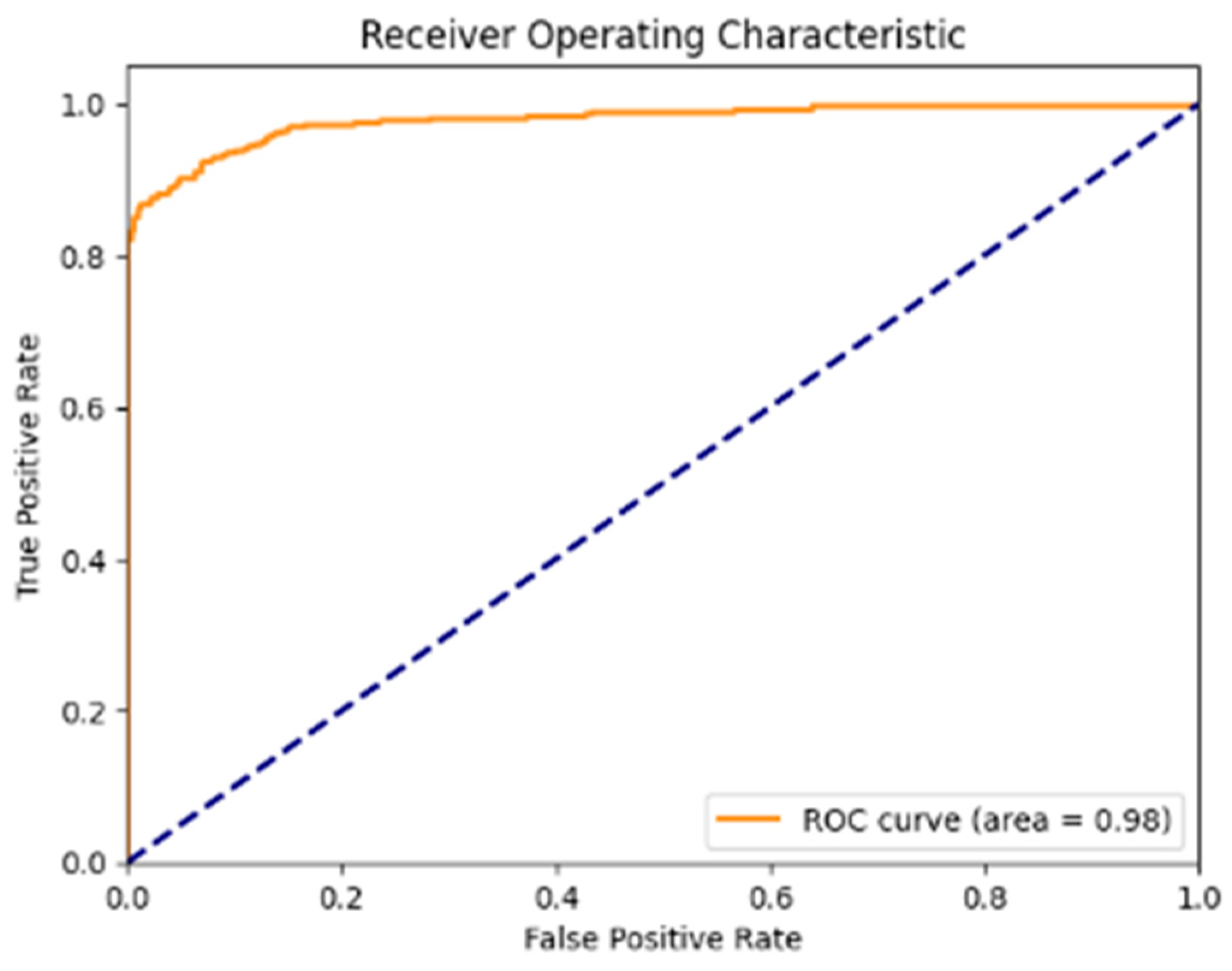
Figure 11 shows that the Receiver Operating Characteristic (ROC) curve is strongly leaning toward the top-left corner, with the ROC-AUC score of 0.9805, which shows the capability of the model to differentiate between malignant and benign lesions with a very low rate of false positives.
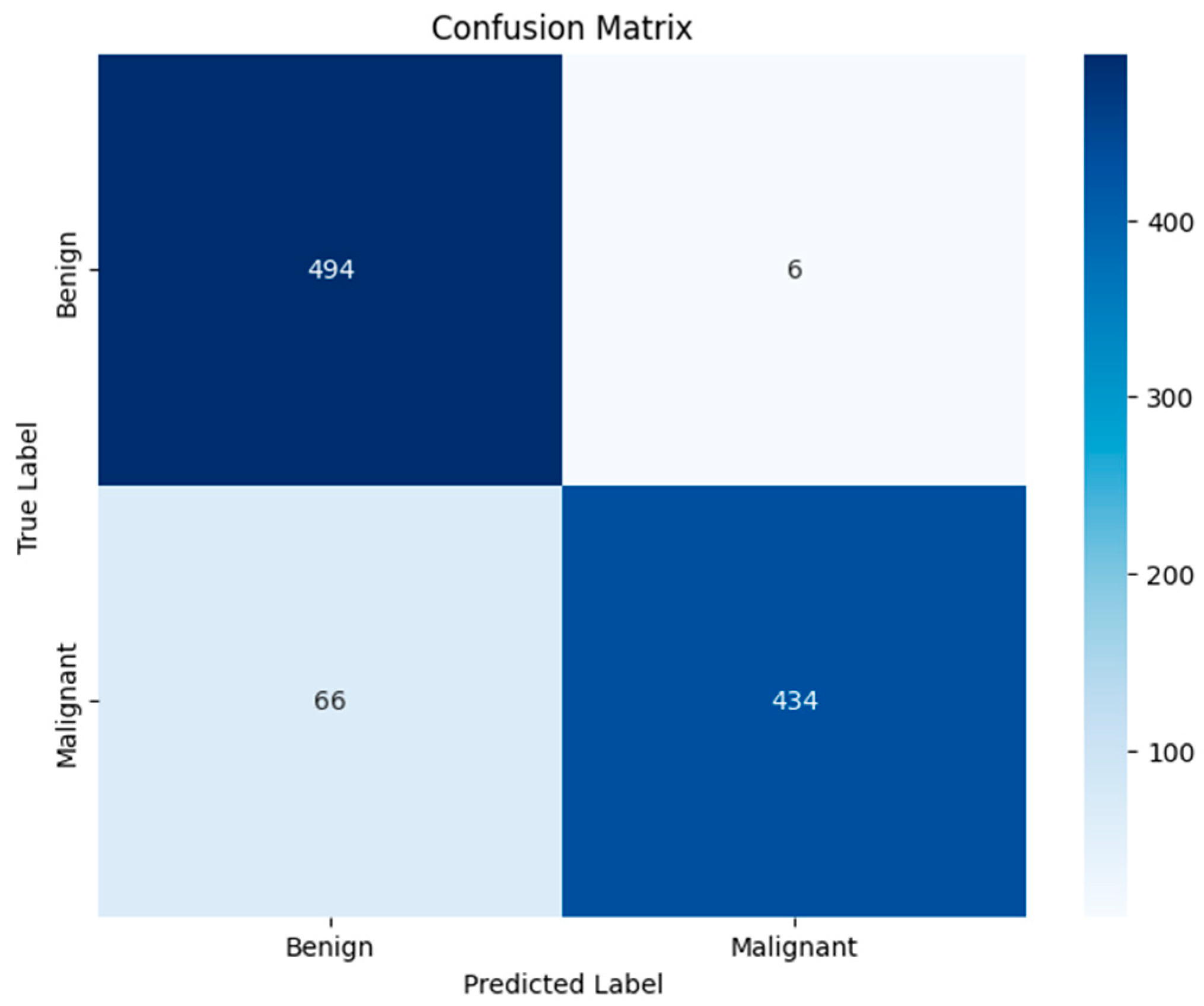
Figure 12 shows the confusion matrix, which shows that the model correctly classified 494 non-cancerous images and 434 cancerous images, while it misclassifies 6 non-cancerous and 69 cancerous images. This shows the model’s strong performance in differentiating between the benign and malignant dermoscopic images, with a high number of true positive and true negative cases. However, in terms of images with cancer, the number of misclassified images is relatively higher, with areas for improvement.
The quantitative results of the proposed model from our study, along with the previous studies, are shown in
Table 1. Our model obtained an accuracy of 0.9280, a precision score of 0.9864, a recall score of 0.8680, and an f1-score of 0.9234. Our model showed the best score in terms of accuracy, precision, and F1 by a small margin, and showed a slightly lesser score in terms of recall, which indicates that the model was accurately classifying the benign images, but was not performing as well in malignant cases.
7. Discussion
In this study, we explored the application of deep learning techniques for skin cancer classification, using the U-Net segmentation, and transfer learning, using the ResNet-50 pre-trained network as the backbone of the feedforward network. We implemented different image pre-processing and augmentation techniques to achieve a robust classification model, from which we achieved an accuracy of 0.9280, precision of 0.9864, recall of 0.8680, and an F1-score of 0.9234 which shows the ability of this approach in skin cancer classification
We implemented U-Net for the segmentation of the lesion area, which enabled the classification model to better focus on the lesion boundaries. We used the segmented image to trace the boundary around the lesion area (as shown in
Figure 10), which highlights the potential of our approach in a classification where it is crucial to detect the lesion boundary. From the confusion matrix (
Figure 12) we can see that we have obtained high true positive and true negative rates, which further confirms our model’s reliability. However, the recall value of 86.80% highlights that there is still room for improvement in the model’s ability to detect the malignant cases effectively in order to identify the harder cancerous lesions.
The accuracy of our model is greater than in the other studies using Kaggle datasets ([
5], [
6]) which have reported accuracies of 0.8424 and 0.8909, respectively. They have not reported other metrics, which makes the comparison challenging. We have used different pre-processing steps, implemented segmentation as a part of pre-processing so that the model focused more on the lesion area, and updated the hyper-parameters to improve the model’s performance. Because of this, our model has achieved a very good performance in terms of precision and accuracy, whereas the recall metric (86.80) indicated room for improvement in malignant case identification.
Our model has shown promising results and also pointed towards room for improvement. We could explore the use of more advanced segmentation models to capture more subtle features. ISIC-2016 [
9] for segmentation and the Kaggle dataset [
10] have provided a strong foundation, and in the future we could also conduct experiments with larger and more diverse datasets, which may improve the generalization of the model.
We have developed our model to be robust and accurate, but there are several real-world challenges that need to be considered before its deployment. Some of the aspects we need to consider are:
Skin types: In clinical practice, it is vital that the model generalizes extremely well and without bias. We acknowledge that the dataset we have used may not fully capture the wide range of skin types we encounter in the real world.
Variation in imaging devices: The images vary significantly depending on the device used to capture them. We have used pre-processing steps to tackle this, but we accept that the significant differences in image quality can affect the model’s ability.
Explainability: Our model classified the images as cancerous or non-cancerous, but did not explain the result, which is important for gaining trust from clinicians. This may hinder the adoption of the model in clinical settings.
Author Contributions
Conceptualization, R.K. and S.G.C.; methodology, R.K. and S.G.C.; software, R.K.; validation, R.K. and J.R.; formal analysis, R.K.; investigation, R.K. and J.R.; resources, R.K., J.R. and A.K.; data curation, R.K.; writing—original draft preparation, R.K.; writing—review and editing, R.K., J.R. and A.K.; visualization, R.K.; supervision, J.R. and A.K.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Leiter, U.; Keim, U.; Garbe, C. Epidemiology of Skin Cancer: Update 2019. SpringerLink. 1970. Available online: https://link.springer.com/chapter/10.1007/978-3-030-46227-7_6 (accessed on 19 May 2024).
- Aljanabi, M.; Özok, Y.E.; Rahebi, J.; Abdullah, A.S. Skin Lesion Segmentation Method for Dermoscopy Images Using Artificial Bee Colony Algorithm. Symmetry 2018, 10, 347. [Google Scholar] [CrossRef]
- Gomathi, E.; Jayasheela, M.; Thamarai, M.; Geetha, M. Skin cancer detection using dual optimization based deep learning network. Biomed. Signal Process. Control 2023, 84, 104968. [Google Scholar] [CrossRef]
- Web.archive.org. (n.d.). Wayback Machine. Available online: https://web.archive.org/web/20130318041656/ (accessed on 19 May 2024).
- Ibrahim, A.M.; Elbasheir, M.; Badawi, S.; Mohammed, A.; Alalmin, A.F.M. Skin Cancer Classification Using Transfer Learning by VGG16 Architecture (Case Study on Kaggle Dataset). J. Intell. Learn. Syst. Appl. 2023, 15, 67–75. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Altameem, A.; Nayak, S.R.; Poonia, R.C.; Saudagar, A.K.J. An Enhanced Transfer Learning Based Classification for Diagnosis of Skin Cancer. Diagnostics 2022, 12, 1628. [Google Scholar] [CrossRef] [PubMed]
- Reis, H.C.; Turk, V.; Khoshelham, K.; Kaya, S. InSiNet: A deep convolutional approach to skin cancer detection and segmentation. Med. Biol. Eng. Comput. 2022, 60, 643–662. [Google Scholar] [CrossRef] [PubMed]
- Arabahmadi, M.; Farahbakhsh, R.; Rezazadeh, J. Deep Learning for Smart Healthcare—A Survey on Brain Tumor Detection from Medical Imaging. Sensors 2022, 22, 1960. [Google Scholar] [CrossRef] [PubMed]
- Gutman, D.; Codella, N.C.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin Lesion Analysis toward Melanoma Detection: A Challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Javid, M.H. Melanoma Skin Cancer Dataset of 10000 Images [Data Set]; Kaggle: San Francisco, CA, USA, 2022. [Google Scholar] [CrossRef]
- Sivakumar, M.S.; Leo, L.M.; Gurumekala, T.; Sindhu, V.; Priyadharshini, A.S. Deep learning in skin lesion analysis for malignant melanoma cancer identification. Multimed. Tools Appl. 2023, 83, 17833–17853. [Google Scholar] [CrossRef]
- Abdelhafeez, A.; Mohamed, H.K. Skin cancer detection using neutrosophic c-means and fuzzy c-means clustering algorithms. J. Intell. Syst. Internet Things 2023, 8, 33–42. [Google Scholar] [CrossRef]
- Zafar, M.; Amin, J.; Sharif, M.; Anjum, M.A.; Mallah, G.A.; Kadry, S. DeepLabv3+-Based Segmentation and Best Features Selection Using Slime Mould Algorithm for Multi-Class Skin Lesion Classification. Mathematics 2023, 11, 364. [Google Scholar] [CrossRef]
- O’Shea, K.; Nash, R. An Introduction to Convolutional Neural Networks. arXiv 2015, arXiv:1511.08458. [Google Scholar]
- Suk, H.I. An Introduction to Neural Networks and Deep Learning. In Deep Learning for Medical Image Analysis; Academic Press: Cambridge, MA, USA, 2017; pp. 3–24. [Google Scholar] [CrossRef]
- Ronneberger, O.F.P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef]
- Canny, J.F. Finding Edges and Lines in Images. Available online: http://hdl.handle.net/1721.1/6939 (accessed on 19 August 2024).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).