RE-XswinUnet: Rotary Positional Encoding and Inter-Slice Contextual Connections for Multi-Organ Segmentation
Abstract
1. Introduction
- (1)
- We integrate RoPE into SwinUNet, verifying the feasibility of RoPE in medical image segmentation and enhancing detail modeling capabilities.
- (2)
- A plug-and-play multi-scale skip connections block, XskipNet, is designed to enhance semantic information fusion capabilities.
- (3)
- We design SCAR Block, a novel downsampling module that preserves structural information while improving translational invariance.
- (4)
- Extensive experiments on the Synapse and ACDC datasets demonstrate competitive performance, and ablation experiments confirm the efficacy and generalizability for every component.
2. Materials and Methods
2.1. Overall Architecture
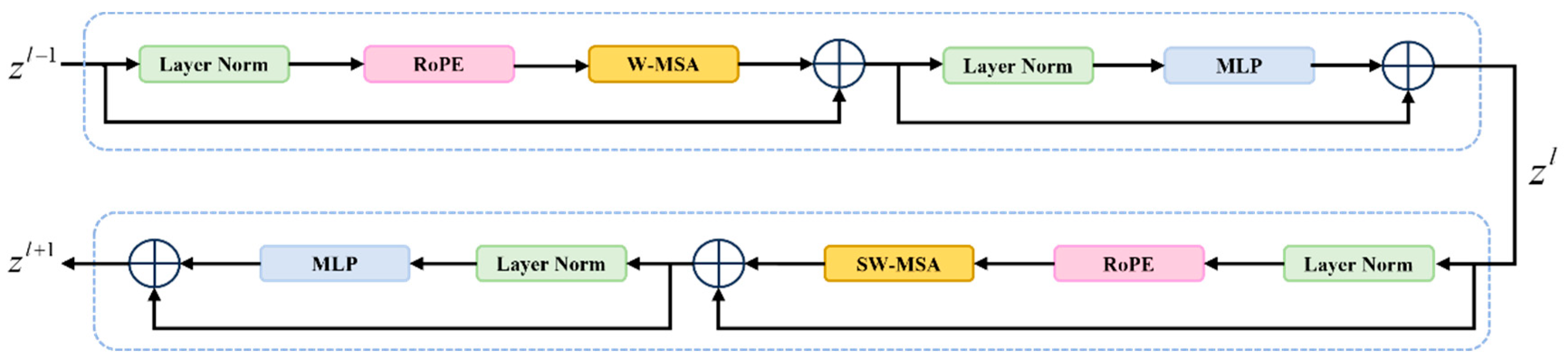
2.2. Rotational Position Embedding
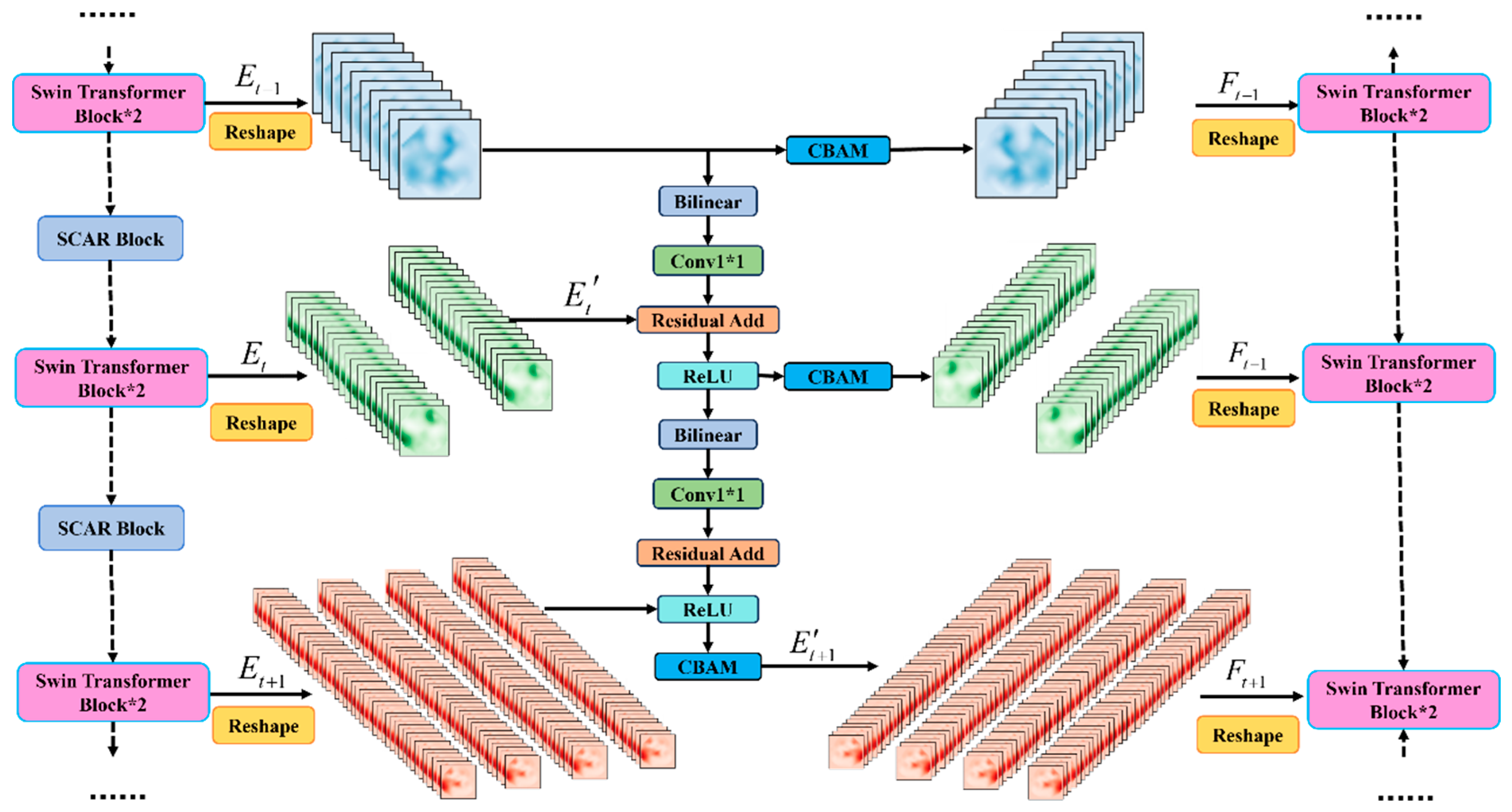
2.3. XskipNet Block
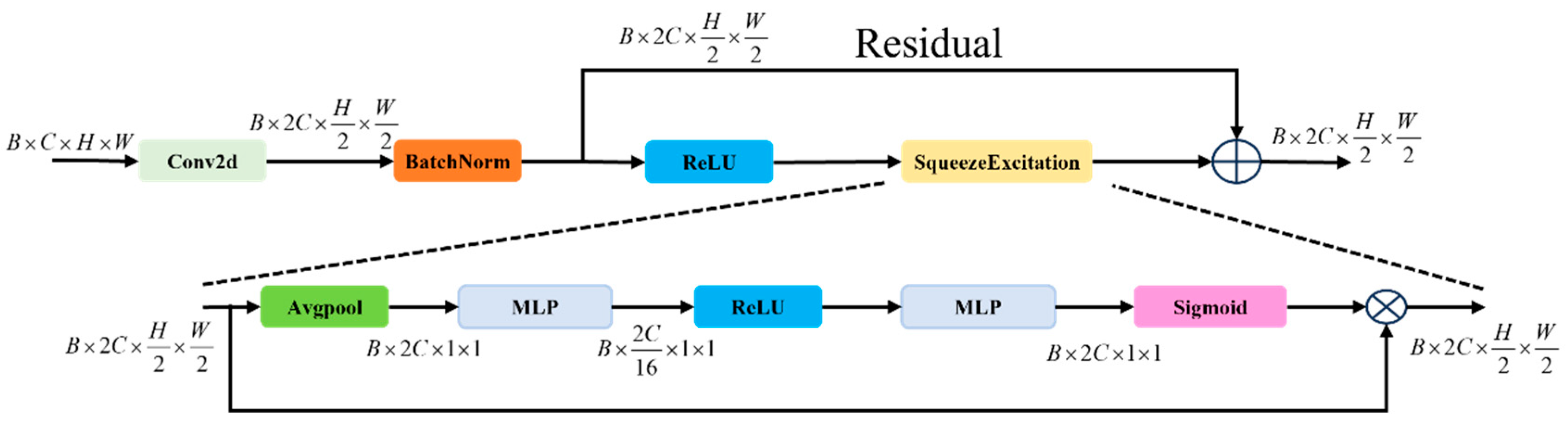
2.4. SCAR Block
3. Results
3.1. Datasets
3.2. Implementation Details
3.3. Evaluation Indicators
3.4. Overall Performance on Synapse Dataset
3.5. Overall Performance on ACDC Dataset
3.6. Model Complexity and Efficiency Analysis
4. Discussion
4.1. Ablation Studies
4.2. Significance Verification Based on t-Test
4.3. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lv, C.; Li, B.; Wang, X.; Cai, P.; Yang, B.; Sun, G.; Yan, J. ECM-TransUNet: Edge-enhanced multi-scale attention and convolutional Mamba for medical image segmentation. Biomed. Signal Process. Control 2025, 107, 107845. [Google Scholar] [CrossRef]
- Xia, Q.; Zheng, H.; Zou, H.; Luo, D.; Tang, H.; Li, L.; Jiang, B. A comprehensive review of deep learning for medical image segmentation. Neurocomputing 2025, 613, 128740. [Google Scholar] [CrossRef]
- Etehadtavakol, M.; Etehadtavakol, M.; Ng, E.Y.K. Enhanced thyroid nodule segmentation through U-Net and VGG16 fusion with feature engineering: A comprehensive study. Comput. Methods Programs Biomed. 2024, 251, 108209. [Google Scholar] [CrossRef] [PubMed]
- Dan, Y.; Jin, W.; Yue, X.; Wang, Z. Enhancing medical image segmentation with a multi-transformer U-Net. PeerJ 2024, 12, e17005. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Jiang, Y.; Peng, Y.; Yuan, F.; Zhang, X.; Wang, J. Medical Image Segmentation: A Comprehensive Review of Deep Learning-Based Methods. Tomography 2025, 11, 52. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 26th International Conference on Neural Information Processing Systems—Volume 1, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Yao, W.; Bai, J.; Liao, W.; Chen, Y.; Liu, M.; Xie, Y. From CNN to Transformer: A Review of Medical Image Segmentation Models. J. Imaging Inform. Med. 2024, 37, 1529–1547. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, J.; Liu, W.; Gao, M.; Hu, X.; Xue, Z.; Liu, Y.; Yan, S. Rwkv-unet: Improving unet with long-range cooperation for effective medical image segmentation. arXiv 2025, arXiv:2501.08458. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Cham, Switzerland, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Du, G.; Cao, X.; Liang, J.; Chen, X.; Zhan, Y. Medical image segmentation based on U-net: A review. J. Imaging Sci. Technol. 2020, 64, 1. [Google Scholar] [CrossRef]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical image segmentation review: The success of u-net. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10076–10095. [Google Scholar] [CrossRef]
- Huang, L.; Miron, A.; Hone, K.; Li, Y. Segmenting Medical Images: From UNet to Res-UNet and nnUNet. In Proceedings of the 2024 IEEE 37th International Symposium on Computer-Based Medical Systems (CBMS), Guadalajara, Mexico, 26–28 June 2024; pp. 483–489. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016, Cham, Switzerland, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Oktay, O.; Schlemper, J.; Le Folgoc, L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the International Workshop on Deep Learning in Medical Image Analysis, Granada, Spain, 20 September 2018; pp. 3–11. [Google Scholar]
- Qi, Y.; Cai, J.; Chen, R. AO-TransUNet: A multi-attention optimization network for COVID-19 and medical image segmentation. Digit. Signal Process. 2025, 164, 105264. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C. ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm. Remote Sens. 2020, 162, 94–114. [Google Scholar] [CrossRef]
- Cai, S.; Tian, Y.; Lui, H.; Zeng, H.; Wu, Y.; Chen, G. Dense-UNet: A novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network. Quant Imaging Med. Surg. 2020, 10, 1275. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.S. Advancements in medical image segmentation: A review of transformer models. Comput. Electr. Eng. 2025, 123, 110099. [Google Scholar] [CrossRef]
- Ashish, V.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual Event, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Krishna, M.S.; Machado, P.; Otuka, R.I.; Yahaya, S.W.; Neves dos Santos, F.; Ihianle, I.K. Plant Leaf Disease Detection Using Deep Learning: A Multi-Dataset Approach. J 2025, 8, 4. [Google Scholar] [CrossRef]
- Pu, Q.; Xi, Z.; Yin, S.; Zhao, Z.; Zhao, L. Advantages of transformer and its application for medical image segmentation: A survey. Biomed. Eng. Online 2024, 23, 14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kaul, C.; Wang, J.; Anagnostopoulos, C.; Murray-Smith, R.; Deligianni, F. Optimizing vision transformers for medical image segmentation. In Proceedings of the ICASSP 2023-2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes, Greece, 4–10 June 2023; pp. 1–5. [Google Scholar]
- Wang, Y.; Qiu, Y.; Cheng, P.; Zhang, J. Hybrid CNN-transformer features for visual place recognition. IEEE Trans. Circuits Syst. Video Technol. 2022, 33, 1109–1122. [Google Scholar] [CrossRef]
- Qiao, X.; Yan, Q.; Huang, W.; Sensing, R. Hybrid CNN-Transformer Network With a Weighted MSE Loss for Global Sea Surface Wind Speed Retrieval From GNSS-R Data. IEEE Trans. Geosci. Remote Sens. 2025, 63, 1–13. [Google Scholar] [CrossRef]
- Bashir, T.; Wang, H.; Tahir, M.; Zhang, Y. Wind and solar power forecasting based on hybrid CNN-ABiLSTM, CNN-transformer-MLP models. Renew. Energy 2025, 239, 122055. [Google Scholar] [CrossRef]
- Tang, H.; Chen, Y.; Wang, T.; Zhou, Y.; Zhao, L.; Gao, Q.; Du, M.; Tan, T.; Zhang, X.; Tong, T. HTC-Net: A hybrid CNN-transformer framework for medical image segmentation. Biomed. Signal Process. Control. 2024, 88, 105605. [Google Scholar] [CrossRef]
- Zheng, S.; Lu, J.; Zhao, H.; Zhu, X.; Luo, Z.; Wang, Y.; Fu, Y.; Feng, J.; Xiang, T.; Torr, P.H. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual Event, 19–25 June 2021; pp. 6881–6890. [Google Scholar]
- Xie, Y.; Zhang, J.; Shen, C.; Xia, Y. Cotr: Efficiently bridging cnn and transformer for 3d medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; pp. 171–180. [Google Scholar]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. Transbts: Multimodal brain tumor segmentation using transformer. In Proceedings of the International conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; pp. 109–119. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; pp. 205–218. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Kumar, S.; Kumar, R.V.; Ranjith, V.; Jeevakala, S.; Varun, S.S.J.C.; Engineering, E. Grey Wolf optimized SwinUNet based transformer framework for liver segmentation from CT images. Comput. Electr. Eng. 2024, 117, 109248. [Google Scholar] [CrossRef]
- Su, J.; Ahmed, M.; Lu, Y.; Pan, S.; Bo, W.; Liu, Y. Roformer: Enhanced transformer with rotary position embedding. Neurocomputing 2024, 568, 127063. [Google Scholar] [CrossRef]
- Shaw, P.; Uszkoreit, J.; Vaswani, A. Self-attention with relative position representations. arXiv 2018, arXiv:1803.02155. [Google Scholar] [CrossRef]
- Li, X.; Cheng, Y.; Fang, Y.; Liang, H.; Xu, S. 2DSegFormer: 2-D Transformer Model for Semantic Segmentation on Aerial Images. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–13. [Google Scholar] [CrossRef]
- Wu, K.; Peng, H.; Chen, M.; Fu, J.; Chao, H. Rethinking and improving relative position encoding for vision transformer. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10033–10041. [Google Scholar]
- Liutkus, A.; Cıfka, O.; Wu, S.-L.; Simsekli, U.; Yang, Y.-H.; Richard, G. Relative positional encoding for transformers with linear complexity. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 7067–7079. [Google Scholar]
- Heo, B.; Park, S.; Han, D.; Yun, S. Rotary position embedding for vision transformer. In Proceedings of the European Conference on Computer Vision, Milan, Italy, 29 September–4 October 2024; pp. 289–305. [Google Scholar]
- Wang, P.; Yang, Q.; He, Z.; Yuan, Y. Vision transformers in multi-modal brain tumor MRI segmentation: A review. Meta-Radiology 2023, 1, 100004. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Li, Y.; Zou, Y.; He, X.; Xu, Q.; Liu, M.; Jin, S.; Zhang, Q.; He, M.M.; Zhang, J. HFA-UNet: Hybrid and full attention UNet for thyroid nodule segmentation. Knowl.-Based Syst. 2025, 328, 114245. [Google Scholar] [CrossRef]
- Sun, P.; Wu, J.; Zhao, Z.; Gao, H. ACMS-TransNet: Polyp Segmentation Network Based on Adaptive Convolution and Multi-Scale Global Context. IAENG Int. J. Comput. Sci. 2025, 52, 474–483. [Google Scholar]
- Zhu, Y.; Zhang, D.; Lin, Y.; Feng, Y.; Tang, J. Merging Context Clustering with Visual State Space Models for Medical Image Segmentation. Qeios 2025, 44, 2131–2142. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, L.; Cheng, S.; Li, Y. SwinSUNet: Pure transformer network for remote sensing image change detection. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–13. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Segmentation Outside the Cranial Vault Challenge. 2015. Available online: https://repo-prod.prod.sagebase.org/repo/v1/doi/locate?id=syn3193805&type=ENTITY (accessed on 17 September 2024). [CrossRef]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Lu, Y.; Wang, Y.; Zhou, Y.; Shen, W.; Fishman, E.; Yuille, A. Domain adaptive relational reasoning for 3d multi-organ segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 656–666. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Jha, A.; Kumar, A.; Pande, S.; Banerjee, B.; Chaudhuri, S. Mt-unet: A novel u-net based multi-task architecture for visual scene understanding. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Virtual, 25–28 October 2020; pp. 2191–2195. [Google Scholar]
- Yu, J.; Qin, J.; Xiang, J.; He, X.; Zhang, W.; Zhao, W. Trans-UNeter: A new Decoder of TransUNet for Medical Image Segmentation. In Proceedings of the 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Istanbul, Turkey, 5–8 December 2023; pp. 2338–2341. [Google Scholar]

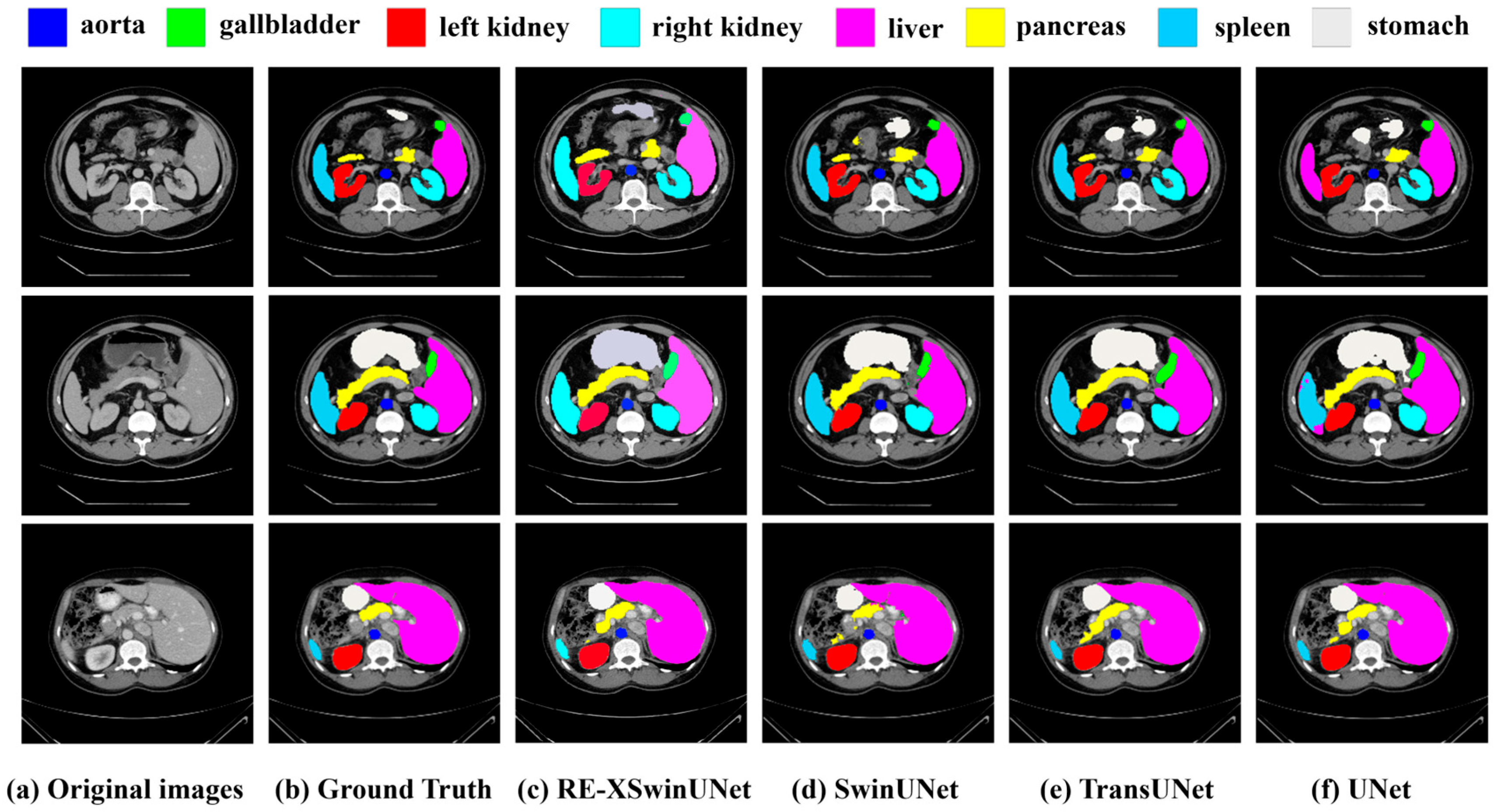
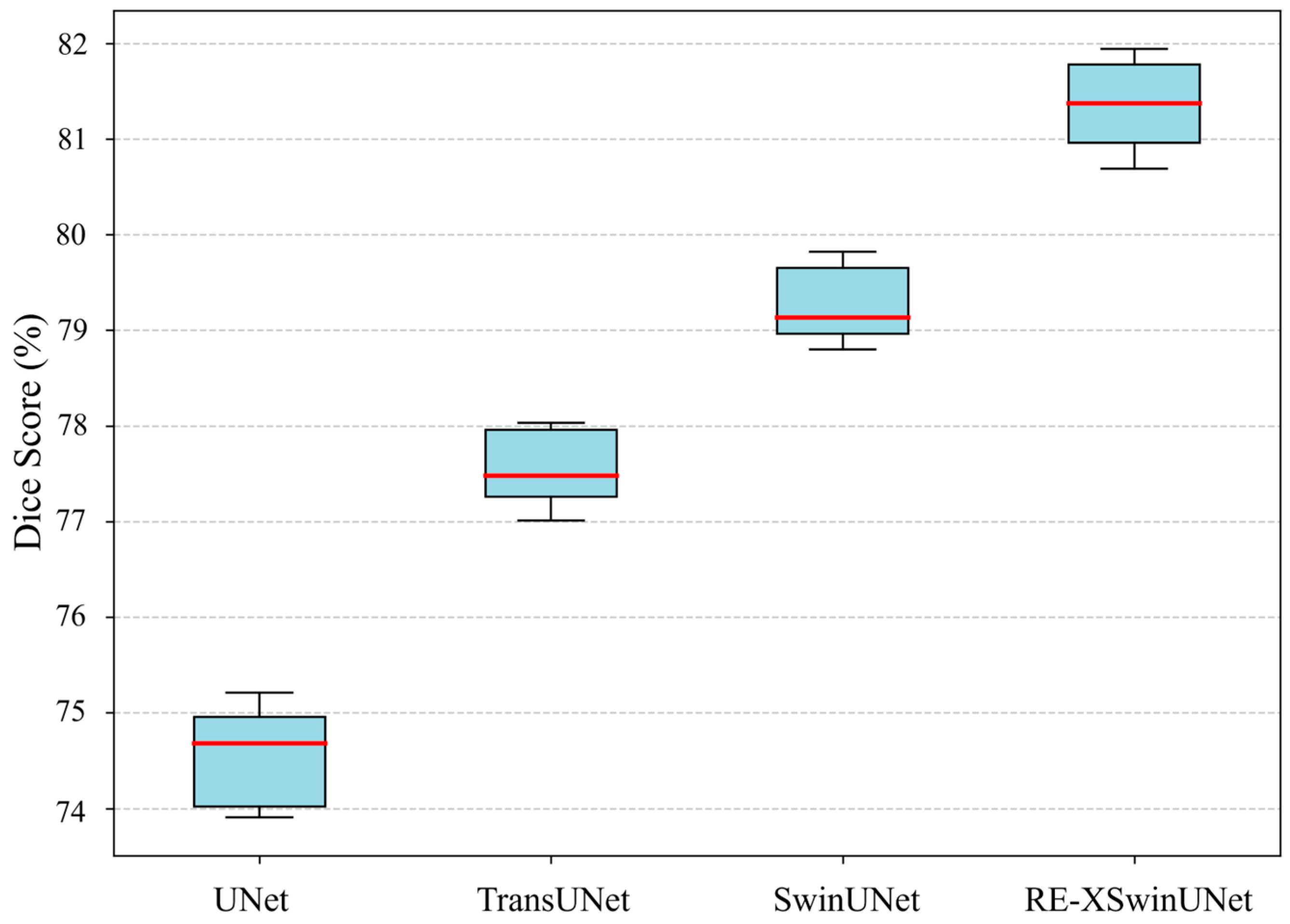
| Method | DSC | HD | Aorta | Gall. | Kidney(L) | Kidney(R) | Liver | Pancreas | Spleen | Stomach |
|---|---|---|---|---|---|---|---|---|---|---|
| V-Net [13,16] | 68.81 | - | 75.34 | 51.87 | 77.10 | 80.75 | 87.84 | 40.05 | 80.56 | 56.98 |
| DARR [16,54] | 69.77 | - | 74.74 | 53.77 | 72.31 | 73.24 | 94.08 | 54.18 | 89.90 | 45.96 |
| U-Net [9,16] | 74.68 | 36.87 | 87.74 | 63.66 | 80.60 | 78.19 | 93.74 | 56.90 | 85.87 | 74.16 |
| AttnUNet [15,16] | 75.57 | 36.97 | 55.92 | 63.91 | 79.20 | 72.71 | 93.56 | 49.37 | 87.19 | 74.95 |
| R50-ViT [16,55] | 71.29 | 32.87 | 73.73 | 55.13 | 75.80 | 72.20 | 91.51 | 45.99 | 81.99 | 73.95 |
| TransUNet [16] | 77.48 | 31.69 | 87.23 | 63.13 | 81.87 | 77.02 | 94.08 | 55.86 | 85.08 | 75.62 |
| CoT-TransUNet [36] | 78.24 | 23.75 | 88.69 | 62.56 | 88.33 | 76.91 | 94.57 | 55.23 | 86.35 | 78.28 |
| MT-UNet [56] | 78.59 | 26.59 | 87.92 | 64.99 | 81.47 | 77.29 | 93.06 | 59.46 | 87.75 | 76.81 |
| SwinUNet [36] | 79.13 | 21.55 | 85.47 | 66.53 | 83.28 | 79.61 | 94.29 | 56.58 | 90.66 | 76.60 |
| RE-XSwinUNet | 81.78 | 18.83 | 87.99 | 68.19 | 83.76 | 81.43 | 94.55 | 65.11 | 91.72 | 81.52 |
| Methods | DSC | RV | Myo | LV |
|---|---|---|---|---|
| R50 U-Net [16,57] | 87.55 | 87.10 | 80.63 | 94.92 |
| R50 Att-UNet [36,57] | 86.75 | 87.58 | 79.20 | 93.47 |
| R50 ViT [36,57] | 87.57 | 86.07 | 81.88 | 94.75 |
| TransUNet [16,57] | 89.71 | 88.86 | 84.53 | 95.73 |
| SwinUNet [36,57] | 90.00 | 88.55 | 85.62 | 95.83 |
| RE-XSwinUNet | 90.95 | 91.24 | 85.52 | 96.08 |
| Methods | Total Params (M) | Estimated Total Size (MB) |
|---|---|---|
| UNet | 31.037 | 567.90 |
| TransUNet | 108.596 | 841.84 |
| SwinUNet | 37.169 | 408.04 |
| RE-XSwinUNet | 40.241 | 423.98 |
| Block | DSC | Aorta | Gallb. | Kidney (L) | Kidney (R) | Liver | Pancreas | Spleen | Stomach |
|---|---|---|---|---|---|---|---|---|---|
| SwinUNet [36] | 79.13 | 85.47 | 66.53 | 83.28 | 79.61 | 94.29 | 56.58 | 90.66 | 76.60 |
| RoPE | 80.38 | 86.32 | 67.78 | 82.76 | 78.50 | 94.12 | 63.43 | 91.19 | 78.92 |
| XskipNet | 79.97 | 85.55 | 67.83 | 82.37 | 78.14 | 94.58 | 62.26 | 91.72 | 77.28 |
| SCAR Block | 80.12 | 86.82 | 66.34 | 84.47 | 81.76 | 94.38 | 56.10 | 91.79 | 79.32 |
| RoPE+XskipNet | 81.15 | 87.25 | 68.05 | 83.25 | 80.35 | 94.52 | 64.85 | 91.45 | 80.28 |
| RoPE+SCAR Block | 80.95 | 87.42 | 67.45 | 84.12 | 81.25 | 94.45 | 62.75 | 91.68 | 80.65 |
| XskipNet+SCAR Block | 80.65 | 86.95 | 67.28 | 84.35 | 81.08 | 94.62 | 60.85 | 91.85 | 79.95 |
| RE-XSwinUNet (All) | 81.78 | 87.99 | 68.19 | 83.76 | 81.43 | 94.55 | 65.11 | 91.72 | 81.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Yang, C.; Yang, D.; Hang, X.; Liu, W. RE-XswinUnet: Rotary Positional Encoding and Inter-Slice Contextual Connections for Multi-Organ Segmentation. Big Data Cogn. Comput. 2025, 9, 274. https://doi.org/10.3390/bdcc9110274
Yang H, Yang C, Yang D, Hang X, Liu W. RE-XswinUnet: Rotary Positional Encoding and Inter-Slice Contextual Connections for Multi-Organ Segmentation. Big Data and Cognitive Computing. 2025; 9(11):274. https://doi.org/10.3390/bdcc9110274
Chicago/Turabian StyleYang, Hang, Chuanghua Yang, Dan Yang, Xiaojing Hang, and Wu Liu. 2025. "RE-XswinUnet: Rotary Positional Encoding and Inter-Slice Contextual Connections for Multi-Organ Segmentation" Big Data and Cognitive Computing 9, no. 11: 274. https://doi.org/10.3390/bdcc9110274
APA StyleYang, H., Yang, C., Yang, D., Hang, X., & Liu, W. (2025). RE-XswinUnet: Rotary Positional Encoding and Inter-Slice Contextual Connections for Multi-Organ Segmentation. Big Data and Cognitive Computing, 9(11), 274. https://doi.org/10.3390/bdcc9110274





