Abstract
Since the beginning of the COVID-19 pandemic almost two years ago, there have been more than 700,000 scientific papers published on the subject. An individual researcher cannot possibly get acquainted with such a huge text corpus and, therefore, some help from artificial intelligence (AI) is highly needed. We propose the AI-based tool to help researchers navigate the medical papers collections in a meaningful way and extract some knowledge from scientific COVID-19 papers. The main idea of our approach is to get as much semi-structured information from text corpus as possible, using named entity recognition (NER) with a model called PubMedBERT and Text Analytics for Health service, then store the data into NoSQL database for further fast processing and insights generation. Additionally, the contexts in which the entities were used (neutral or negative) are determined. Application of NLP and text-based emotion detection (TBED) methods to COVID-19 text corpus allows us to gain insights on important issues of diagnosis and treatment (such as changes in medical treatment over time, joint treatment strategies using several medications, and the connection between signs and symptoms of coronavirus, etc.).
1. Introduction
Automatic scientific paper analysis is a fast-growing area of study. In recent years, there has been huge progress in the field of natural language processing (NLP), and very powerful neural network language models have been trained. In the area of NLP, the following tasks are typically considered: text classification/intent recognition—classification of text pieces into a number of categories [1]; sentiment analysis—estimating how positive or negative the text is [2]; named entity recognition (NER)—extraction of named entities from text (for example, names of medicines, or diagnoses), and determination of their type; keyword extraction [3]—a task similar to NER; text summarization–generation of a short version of the original text, or selection of the most important text pieces [4]; question answering—finding the exact answer to the question from text [5]; open-domain question answering (ODQA)—finding the exact answer to the question somewhere in the large text corpus [6].
Since the beginning of COVID-19 pandemic almost two years ago, there have been more than 700,000 scientific papers published on the subject. An individual researcher cannot effectively work with such a huge text corpus and, therefore, some help from AI is highly needed. Thus, at the very beginning of COVID-19 pandemic, a research challenge has been launched on Kaggle to analyze scientific papers on the subject. The dataset behind this competition is called CORD, and it contains a constantly updated corpus of everything that is published on topics related to COVID-19 [7]. Currently, it contains about 700,000 scholarly articles, over half of them with full text, about COVID-19, SARS-CoV-2, and related coronaviruses. This freely available dataset is provided to the global research community to apply recent advances in natural language processing and other AI techniques to generate new insights in support of the ongoing fight against the disease.
The rich text corpus of COVID-19 papers has attracted the attention of the scientific community to develop data-driven methods and discover insightful knowledge. Applications of different NLP methods to scientific literature have been covered in this article [8]. The idea to use AI to extract meaningful insights from scientific papers, especially the CORD dataset, has been previously explored and is still continuing. The CORD dataset has quickly generated a number of data science and computing applications [9]. The solutions cover topics from information retrieval to NLP, including applications in document search [10], question answering [11], and text summarization. Resources for text mining researchers and practitioners include pretrained COVID-19 domain language models, knowledge graphs, and embeddings [12].
For solving NER problems, pretrained contextual language models are used in modern text mining systems. These models are state-of-the-art in NLP and have significantly outperformed previous baselines on the full spectrum of language-based tasks [12]. Many works in the text mining field leverage domain-adapted BERT [13] models such as SciBERT [14] and BioBERT [15], which have been fine-tuned to scientific and biomedical texts, respectively. PubMedBERT [16] is the closest model of this kind in the medical domain, and it uses a specially adapted tokenizer and pre-trained BERT, fine-tuned on texts from the PubMed repository [17].
Knowledge graphs provide a model of entities and relationships in a particular domain. These graphs can be used to represent background knowledge and can also be used to infer or discover new relationships through reasoning. Several COVID-19 knowledge graphs have been constructed by combining relations detected in the literature with other ontologies and databases of structured relationships [12]. The largest one is CovidGraph [18], which combines literature, case statistics, genomic and molecular data. The Knowledge Graph Toolkit [19] integrates the CORD-19 corpus with gene, chemical, disease, and taxonomic information from Wikidata [20]. Knowledge graphs can also support automated reasoning, inference, and the potential discovery of novel relationships.
Embeddings are computed vector representations of spans of text that capture semantic and syntactic similarities between them. There are lots of different embedding methodologies, computing embeddings at different levels of granularity, for word tokens, named entities, sentences, paragraphs, and documents. Paper and concept embeddings have been used by several systems to support search and retrieval over the COVID-19 literature [12]. The SPECTER embedding method computes paper embeddings using a SciBERT model [14] pretrained on relatedness signals derived from the citation graph [21]. SPECTER paper embeddings have been shown to successfully capture paper similarity and are available for all papers in CORD-19. Also available for papers in CORD-19 are clinical concept embeddings trained using the Jet algorithm [22], relation embeddings trained using SeVeN [23] and network co-occurrence embeddings [24] for biomedical entities computed using CORD-19-on-FHIR. Embeddings can be used to retrieve similar texts; for example, to find an answer to a query, we can use the embedding of that query and find all documents from the same embedding space that are close in terms of cosine similarity.
In our paper, we present an architecture of a system that derives specific insights from the corpus of COVID-19 papers (CORD) by applying NLP methods based on PubMedBERT model. We also use entity mapping to standard medical ontologies, such as Unified Medical Language System (UMLS) [25], for better entity categorization and some knowledge graph inference. The proposed architecture uses NoSQL database to store entity-relation metadata, which allows us to use DBMS SQL-based querying to perform semantically rich queries over text corpus. Another novelty of the proposed system is to apply text-based emotion detection (TBED) and knowledge graphs to show changes in medical treatment over time and joint treatment strategies using several medications.
2. Materials and Methods
The main idea of the approach is to extract as much semi-structured information from the text as possible and then store it into a NoSQL database for further querying and processing. Storing information in the database would allow us to make some very specific queries to answer some of the questions, as well as to provide a visual exploration tool for medical experts for structured search and insight generation. The overall architecture of the proposed system is shown in Figure 1.
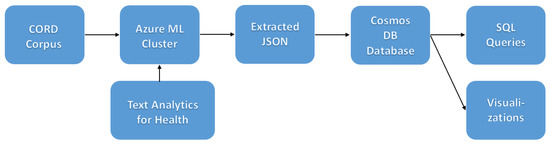
Figure 1.
The architecture of the proposed system for scientific texts analysis.
In the practical implementation of the system, we used different Microsoft Azure technologies, such as Text Analytics for Health [26] and Cosmos DB [27]. The Python programming language developed under an OSI-approved open source license [28] was used throughout the project to make some custom data manipulations and visualizations. For visualization of connection graphs, we used Gephi, the open graph viz platform [29].
2.1. COVID-19 Scientific Papers and CORD Dataset
We analyze the public dataset with scientific papers on topics related to COVID-19 (CORD dataset) [7], which contains more than 700,000 scientific papers, about half of them with the full text [8]. The structure of the dataset is the following:
- Metadata file (“Metadata.csv”) contains the most important information for all publications in one place. Each paper in this table has an identifier “cord_uid”. This identifier is not completely unique throughout the table, but can mostly be used to identify the paper. Other information in metadata.csv includes:
- ○
- Title of publication,
- ○
- Journal,
- ○
- Authors,
- ○
- Abstract,
- ○
- Data of publication,
- ○
- DOI.
- Full-text papers in the “document_parses” directory, in the form of structured text in JSON format.
- Pre-built document embeddings that map cord_uid-s to float vectors that reflect the overall semantics of the paper.
For our research, we focus on paper titles and abstracts because they contain the most important and compressed information from the paper.
2.2. Text Analytics
To make some insights from text, we performed Named Entity Recognition on paper abstracts. Having obtained specific entities that are present in text, we could then perform a semantically rich search of the text that answers specific questions, as well as obtaining data on the co-occurrence of different entities, figuring out specific scenarios or treatment. For this purpose, we use a cognitive service called Text Analytics for Health [12], which is based on pretrained PubMedBERT language model [16] packaged and exposed as a complete ready-to-use REST service that can:
- Perform basic NER on medical and near-medical terms and return the list of entities;
- Do entity mapping to standard medical ontologies, such as Unified Medical Language System (UMLS) [25];
- Extract relations between entities inside the text, such as «TimeOfCondition», etc.;
- Detect negation, which indicates that an entity was used in a negative context, for example, “COVID-19 diagnosis did not occur”.
To perform the analysis of CORD documents, we used Text Analytics Python SDK [30], as well as direct REST calls to Text Analytics service to get JSON document that describes entity/relation data from each paper abstract (see Figure 2a). The structure of this JSON document and the underlying entities/relations are hierarchical and can be effectively stored in a NoSQL database.
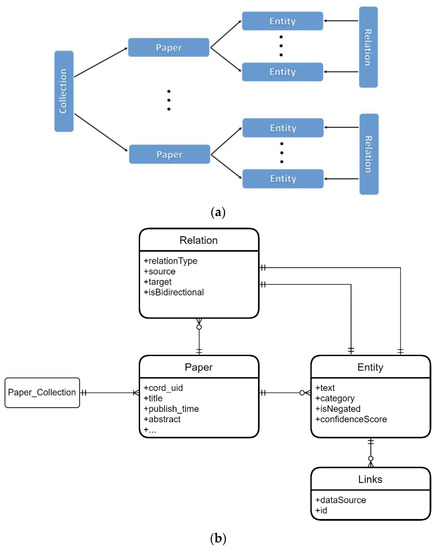
Figure 2.
(a) The overall hierarchical structure of entity/relation metadata for each paper abstract from text collection. Each paper from a collection has a number of entities and corresponding relations between them within the scope of one paper/abstract. (b) Entity diagram corresponding to Cosmos DB document store representing papers/entities/relations. Because we do not have relations between entities in different papers, we can store each paper metadata as one separate JSON document, together with entities and relations between entities.
To speed up data processing for thousands of papers, we used Azure Machine Learning [31]. One of the functions of Azure ML is scheduling jobs (in the form of arbitrary Python scripts) to run on compute clusters with a predefined environment. We used the so-called parallel sweep job, which schedules a number of experiments to run in parallel, which is often used for hyperparameter model optimization and parallel training. In our case, a cluster of N = 8 low-power D3 CPU machines have been used, and «number» parameter for a sweep job that took values from 0 to N-1. Inside the processing loop, we took only rows with those numbers from the original papers’ dataset that gave the remainder equal to «number» when divided by N. Processing results from all machines on the cluster were directly stored in Cosmos DB NoSQL database.
2.3. Entity/Relation Data Storage
As a result of parallel paper processing, a collection of JSON documents was obtained— one for each paper—containing extracted entities and relations, as shown in Figure 2a. This structure is hierarchical, and the best way to store and query it is to use NoSQL approach to data storage. We used Cosmos DB SQL [27], a part of the universal Cosmos DB database that can store and query semi-structured data like our JSON collection. In the collection, entities and relations are stored as one JSON document per paper, and we can represent it by entity/relation diagram in Figure 2b.
With Cosmos SQL, we can formulate semantically rich queries to answer specific questions. For example, to find out all diagnoses connected to a specific medication and corresponding papers, we can use the following query:

2.4. Unified Medical Language System (UMLS)
The Unified Medical Language System (UMLS) [24], initiated in 1986, is a compendium of many controlled vocabularies in the biomedical sciences. It may be viewed as a comprehensive thesaurus and ontology of biomedical concepts.
Text Analytics for Health gives comprehensive ontology mappings for entities where possible, accessible via links field. Each entity may be mapped to several ontologies. In our research, we chose to use one ontology–UMLS–and combine all entities together with their corresponding UMLS ontology IDs. Here is an example of a query that looks for all papers containing certain ontology ID (UMLS ontology ID C0003451 corresponds to “Antiviral Agents “):

2.5. Entity and Relation Types
Text Analytics for Health also gives us classification for entity and relation types. Different entity and relation types understood by the service are presented in Table 1.

Table 1.
Entity and relation types extracted by Text Analytics for Health and the number of their occurrences in the papers dataset.
Relations correspond to short-distance relations inside the text, typically within one sentence, as shown in Figure 3.

Figure 3.
The example of entity/relation metadata text markup. The medications are highlighted in yellow, the dosages are highlighted in green, the frequencies are highlighted in grey.
Note that if we want to have more fine-grained control over entity types, we can employ some additional reasoning based on the UMLS knowledge graph. As an example, if we want to distinguish between “pharmacological substances” and “biological substances” (which are both included in the category “Medications”), we can consider UMLS “Semantic Types” to further categorize entities based on their UMLS ID.
The presence of relations allows us to formulate more semantically rich queries, for example, we can find out which dosages of a specific medication are used in the treatment, together with corresponding papers:

Another important query would be to find out all unique medications mentioned in the collection and group them by UMLS ID. For example, this will unify different ways to call COVID-19 disease (such as SARS-CoV-2) under one group, corresponding to UMLS ID C5203670. The following query retrieves the table of medications with their UMLS IDs:

The same query can be formulated to retrieve other entity types, such as diagnoses and treatments.
In order to perform grouping, as well as some further data processing and visualization, we exported the data from this and similar queries to Pandas DataFrame, which can be further processed and visualized in Python. Cosmos DB supports running of Python-based Jupyter Notebooks inside the Data Explorer, which creates a natural way to query data using the Cosmos DB engine and then do final processing and visualization in Python.
3. Results and Discussion
Using the proposed approach, we managed to get some automatic insights from the corpus of COVID-19 articles. We explored the top-mentioned terms, change in numbers of mentions during the year 2020, and the connection between terms’ (co-occurrence) for some types of named entities. The exploration helps structure scientific papers and understand trends in COVID-19 pandemic description and history of pandemic development.
3.1. TOP Mentioned Entities
The results of TOP mentioned entities belonging to the class of medications, medication classes, treatments, symptoms, and diagnoses are presented in Figure 4 and Figure 5. The Red line shows the percentage of articles with entity mentions in a negative context in all articles that mention this entity.
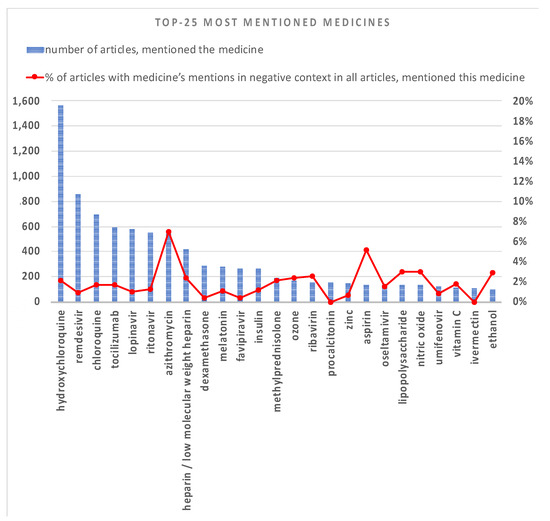
Figure 4.
TOP-25 most mentioned medicines (UMLS Semantic Types: “pharmacological substances” and “biological substances”) in COVID-19 papers from 1 January 2019 till 1 February 2020. The blue histogram, left y-axis corresponds to the number of articles where medicine was mentioned. The red solid line, right y-axis corresponds to the percentage of articles with medicine’s mentions in a negative context with respect to all articles mentioning this medicine.
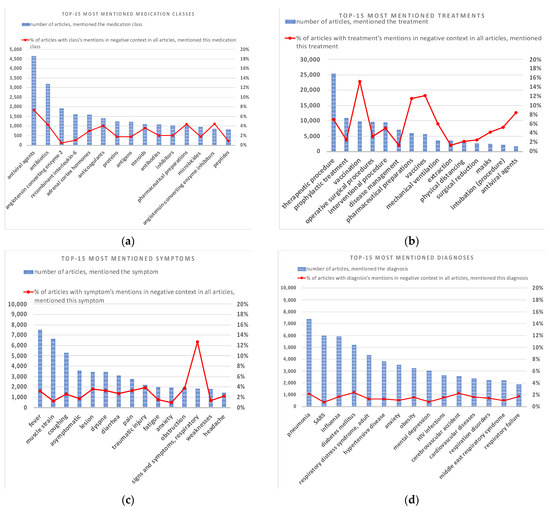
Figure 5.
TOP-15 mentions and percentage of articles with negative mentions (percentage of articles with entity’s mentions in negative context with respect to all articles mentioning this entity). (a) medication classes (including UMLS semantic types: “Pharmacologic Substance”, “Antibiotic”, “Amino Acid, Peptide, or Protein Enzyme”, ”Biologically Active Substance”, ”Immunologic Factor”, “Chemical Viewed Functionally”; (b) treatments (including UMLS semantic types: “Therapeutic or Preventive Procedure”, ”Health Care Activity”, “Pharmacologic Substance”, ”Medical Device”); (c) signs or symptoms (including UMLS semantic groups: “Injuries and Poisonings”, “Pathologic Functions”, “Immunologic Factors”); (d) diagnoses (including UMLS semantic groups: “Disease or Syndrome”, “Mental or Behavioral Dysfunction”).
Figure 4 illustrates TOP-25 most mentioned medications in 2020. They include different types of pharmacological and biological substances. As one can see, the most mentioned medicine is “hydroxychloroquine”. At the beginning of the pandemic, great hopes were pinned on antimalarial drugs (chloroquine, hydroxychloroquine), as well as their combination with azithromycin. “Azithromycin” is also shown as the most negatively mentioned medicine in Figure 4. Other promising drugs were HIV protease inhibitors (such as lopinavir and ritonavir), and they are also included in TOP-6 of medicines. Now, the COVID-19 Treatment Guidelines Panel recommends against the use of chloroquine and hydroxychloroquine and/or azithromycin; lopinavir/ritonavir and other HIV protease inhibitors for the treatment of COVID-19 in hospitalized patients and non-hospitalized patients [32,33].
Figure 5 illustrates the most mentioned medication classes (5a), treatments (5b), symptoms (5c), and diagnoses (5d). To consolidate entity classes, the UMLS semantic types were used.
As one can see in Figure 5a, “antiviral agents “and “antibiotics” appears most often in the CORD dataset. Other medication classes such as “angiotensin-converting enzyme 2”, “recombinant interleukin-6”, “adrenal cortex hormones”, and others have less than 2000 mentions. The percent of negative mentions does not exceed 8 for all of the medication classes, but “antiviral agents” are used in negative context most often. The TOP-5 most mentioned treatments (Figure 5b) include “therapeutic procedures “(over 25,000 mentions), “prophylactic procedures”, “vaccination”, and “operative surgical procedures” (about 10,000 mentions of each). Figure 5b shows that the most frequently mentioned in negative context treatment is «vaccine/vaccination». Checking the articles with negative references suggests that most often “vaccination” is mentioned in abstracts of articles published in 2020 (including in electronic form) in the context: “Currently there is no approved drug or vaccine for the disease” [34] (“Control of this pandemic disease is challenging because there is no effective drug or vaccine available against this virus and this situation demands an urgent need for the development of anti-SARS-CoV-2 potential medicines” [35], “Despite some advances in drug treatments of medical complications in later stages of the disease, the pandemic’s death toll is tragic, since no vaccine or specific antiviral treatment is currently available” [36], etc.). Thus, a large number of negative mentions of «vaccine/vaccination» is associated with the concern of scientists about the lack of a vaccine against COVID-19 at the beginning of the pandemic, and not with the negative effect of the vaccination as such.
The TOP-3 symptoms of COVID-19 (Figure 5c) are “fever” (over 7000 mentions), “muscle strain” (over 6000 mentions), and “coughing” (over 5000 mentions). “Asymptomatic”, “lesion”, and “dyspnea” have about 3500 mentions each. About 13% of mentions in a negative context are related to “respiratory signs and symptoms”, in contexts similar to: “Respiratory signs and symptoms weren’t observed”. Illustrating diagnoses appearing in CORD dataset (Figure 5d), we removed “COVID-19” to show other diseases more clearly. Most often, a diagnosis of «pneumonia» is recorded in a CORD dataset (over 7000 mentions). The percent of negative mentions is lower than 2.5% for all of the diagnoses.
3.2. Treatment Strategy over Time
Another important subject to study is the change of named entity’s mentions over time, as we believe they can indicate changes in treatment strategies (Figure 6). For most frequent medications, we considered the change in mentions over time, shown as the percentage of the entity’s mentions with respect to all articles in the CORD dataset (yellow lines).
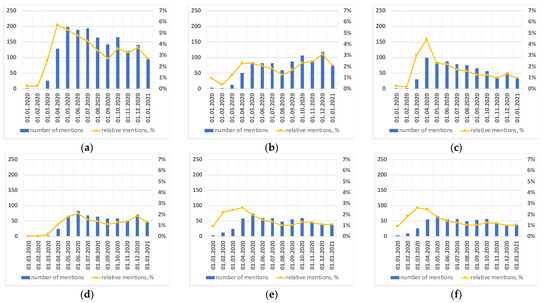
Figure 6.
Number of monthly mentions of medications (blue histogram, left y-axis ) and percentage of mentions w.r.t. all articles (yellow solid line, right y-axis) (a) hydroxychloroquine (UMLS ID C0020336); (b) remdesivir (UMLS ID C4726677); (c) chloroquine (UMLS ID C0008269); (d) tocilizumab (UMLS ID C1609165); (e) lopinavir (UMLS ID C0674432); (f) ritonavir (UMLS ID C0292818).
Considering the graphs of entities’ mentions during the year, we assumed that the peak of research interest matches up the month with the highest number of mentions. There is usually a gap of about six months between receiving and publishing an article; however, most of the journals provided fast track and electronic publishing for COVID-19-related articles in 2020, so our assumption is valid. Figure 6 shows the decrease in mentions of “hydroxychloroquine”/”chloroquine” (Figure 6a,c) and “lopinavir”/”ritonavir” (Figure 6e,f) during the year–the peak of interest was in April 2020, then one can see the steady decline in mentions. There are two peaks of interest for “remdesivir” (April and December 2020–Figure 6b) and “tocilizumab” (June and December 2020–Figure 6d). In the middle of 2020 remdesivir was not recommended for COVID-19 treatment because of insufficient clinical data. But now remdesivir is approved by the Food and Drug Administration (FDA) for the treatment of COVID-19 in hospitalized adult and pediatric patients (aged ≥ 12 years and weighing ≥ 40 kg) [37].
3.3. Terms Co-Occurrence
To observe which terms frequently occur together, we computed co-occurrence matrix, which iI row i and column j contains a number of co-occurrences of terms i and j in the same abstract (one can notice that this matrix is symmetric) and then visualized co-occurrences, using two types of visualizations:
- Sankey diagram allowed us to investigate relations between two types of terms, e.g., diagnosis and treatment (Figure 7a);
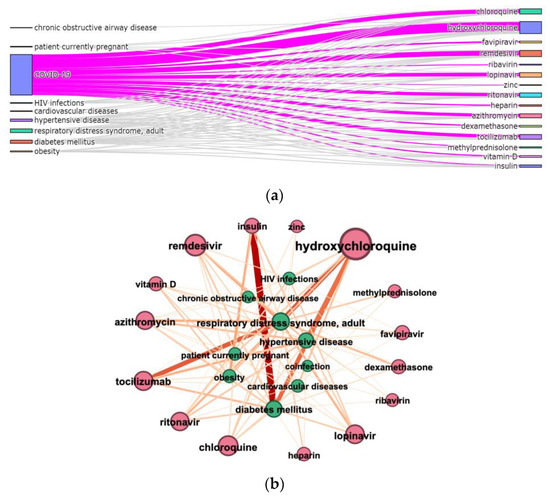
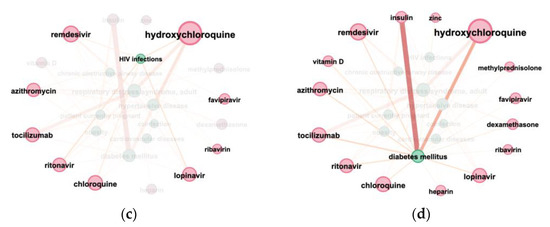 Figure 7. The connection between diagnoses and medicines in COVID-19 articles from 1 January 2019 to 1 February 2020 (a) Sankey diagram: diagnoses and medicines, the diagnoses list includes COVID-19; (b) connection graph: diagnoses and medicines, the diagnoses list excludes COVID-19; (c) connection graph: HIV infections (UMLS ID C0019693) and medicines; (d) connection graph: diabetes mellitus (UMLS ID C0011849) and medicines.
Figure 7. The connection between diagnoses and medicines in COVID-19 articles from 1 January 2019 to 1 February 2020 (a) Sankey diagram: diagnoses and medicines, the diagnoses list includes COVID-19; (b) connection graph: diagnoses and medicines, the diagnoses list excludes COVID-19; (c) connection graph: HIV infections (UMLS ID C0019693) and medicines; (d) connection graph: diabetes mellitus (UMLS ID C0011849) and medicines. - Connection graphs helped us visualize the co-occurrence of terms of the same or different types (e.g., which medications are mentioned together) (Figure 7b–d, Figure 8 and Figure 9).
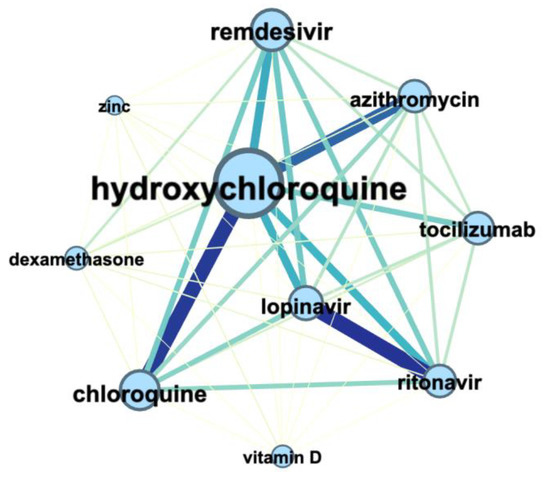 Figure 8. The connection between the medicines (UMLS Semantic Types: “pharmacological substances” and “biological substances”) in COVID-19 articles from 1 January 2019 till 1 February 2020. The node and font size correspond to the number of entities’ mentions; the color and thickness of the edge corresponds to the number of connections between entities.
Figure 8. The connection between the medicines (UMLS Semantic Types: “pharmacological substances” and “biological substances”) in COVID-19 articles from 1 January 2019 till 1 February 2020. The node and font size correspond to the number of entities’ mentions; the color and thickness of the edge corresponds to the number of connections between entities.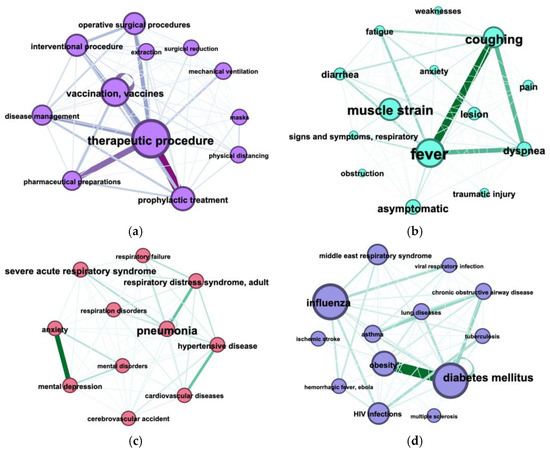 Figure 9. The connection between named entities in COVID-19 articles from 1 January 2019 to 1 February 2020 The node and font size corresponds to the number of entities’ mentions; the color and thickness of the edge correspond to the number of connections between entities. (a) treatments (including UMLS semantic types: “Therapeutic or Preventive Procedure”, ”Health Care Activity”, “Pharmacologic Substance”, ”Medical Device”); (b) signs or symptoms (including UMLS semantic groups: “Injuries and Poisonings”, “Pathologic Functions”, “Immunologic Factors”); (c) diagnoses (including UMLS semantic groups: “Disease or Syndrome”, “Mental or Behavioral Dysfunction”) concomitant COVID-19 (i.e., those that may be caused by a coronavirus infection); (d) diagnoses (including UMLS semantic groups: “Disease or Syndrome”, “Mental or Behavioral Dysfunction”) unrelated directly COVID-19.
Figure 9. The connection between named entities in COVID-19 articles from 1 January 2019 to 1 February 2020 The node and font size corresponds to the number of entities’ mentions; the color and thickness of the edge correspond to the number of connections between entities. (a) treatments (including UMLS semantic types: “Therapeutic or Preventive Procedure”, ”Health Care Activity”, “Pharmacologic Substance”, ”Medical Device”); (b) signs or symptoms (including UMLS semantic groups: “Injuries and Poisonings”, “Pathologic Functions”, “Immunologic Factors”); (c) diagnoses (including UMLS semantic groups: “Disease or Syndrome”, “Mental or Behavioral Dysfunction”) concomitant COVID-19 (i.e., those that may be caused by a coronavirus infection); (d) diagnoses (including UMLS semantic groups: “Disease or Syndrome”, “Mental or Behavioral Dysfunction”) unrelated directly COVID-19.
Graph visualization (Figure 7b–d, Figure 8 and Figure 9) allows us to show the number of entities’ mentions (node and title size) and the number of connections between entities (color and thickness of the edge). To simplify visualization, the diagnosis “COVID-19”, connected with all other entities, has been removed in Figure 7b–d and Figure 9c,d.
The connection of different medications and diagnoses mentioned in CORD dataset is shown in Figure 7b. Figure 7c,d zooms on medicines connected with “HIV infections” and “diabetes mellitus”, respectively. “Diabetes mellitus” is most often connected with “insulin”, while “HIV infections” have no such co-occurrence. This conclusion can be considered as additional evidence of the results’ correctness.
Figure 8 illustrates the connection between the medicines (UMLS Semantic Types: “pharmacological substances” and “biological substances”) in COVID-19 articles and shows the co-occurrence of such combinations as “hydroxychloroquine” + “chloroquine” + “azithromycin” and “lopinavir” + “ritonavir”.
Co-occurrence of COVID-19 treatment methods is illustrated in Figure 9; it shows co-occurrence of treatments (Figure 9a), signs or symptoms (Figure 9b), diagnoses concomitant COVID-19 (Figure 9c), diagnoses unrelated directly COVID-19 (Figure 9d) in COVID-19 articles.
Co-occurrence of COVID-19 treatment methods, illustrated in Figure 9a, shows a strict connection between “therapeutic procedure” and “therapeutic preparations”, “prophylactic treatment”, “vaccination/vaccines”. So we can see two main directions: treatment and prophylactic of COVID-19. The three most common symptoms occurring together are “fever”, “coughing”, “dyspnea” (Figure 9b), the most occurring together diagnoses are “mental depression” and “anxiety” (Figure 9c), “diabetes mellitus “and “obesity” (Figure 9d).
4. Conclusions
The architecture of a proof-of-concept system for knowledge extraction from large corpora of medical texts was described. PubMedBERT and Text Analytics for Health service were used to perform the main task of extracting entities and relations from text, and then a number of Azure services together to build a query tool for medical scientists and to extract some visual insights. The system integrates NLP, text-based emotion detection (TBED), and knowledge graph methods and applies them to COVID-19 text corpus, allowing us to gain insights into important issues of diagnosis and treatment (such as changes in medical treatment over time, joint treatment strategies using several medications, the connection between signs and symptoms of coronavirus, etc.).
The same approach can be applied to other scientific areas, however, in most cases it will require training a custom neural network model to perform entity extraction and, consecutively, labeling the dataset of entities. If we also want to explore the knowledge graph-based inference, problem domain ontology needs to exist or be constructed for the problem domain in question, and labeling should include mapping of each entity to the corresponding ontology node.
In future research, it could be useful to extend our approach to processing full-text articles as well. In this case, we should consider slightly different criteria for the co-occurrence of terms (e.g., in the same paragraph vs. the same paper). The use of full-text articles also imposes additional distortions in the interpretation of negative mentions, which is why we worked with abstracts during our study. Another fruitful direction of research would be to further look into sentence structure and extract more meaning from subject–verb–object relationships. This much more fine-grained document structure can also be stored in a NoSQL database with a slightly more complex schema and then further correlated with general language ontology to collide synonyms and perform some semantic generalization.
Overall, we believe that the work in the direction of knowledge and insight extraction from natural scientific texts is a key to increasing productivity of a scientist, giving him/her some semantically rich tools to augment the research process.
Author Contributions
Conceptualization, D.S., T.P. and A.G.; methodology, T.P., D.S. and A.G.; data analysis, visualization, and validation, T.P., D.S. and V.S.; writing—original draft preparation, T.P. and D.S.; writing—review and editing, T.P., D.S. and A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Ministry of Science and Higher Education of the Russian Federation (Grant № 075-15-2020-801).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
A tool for knowledge extraction from scientific COVID-19 papers is available online as Python code from the following Github repository: https://github.com/shwars/COVIDPaperAnalysis (accessed on 25 December 2021).
Acknowledgments
The authors thank MSU Institute for Artificial Intelligence for administrative and technical support.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
List of acronyms and abbreviations
| AI | artificial intelligence |
| BERT | Bidirectional Encoder Representations from Transformers |
| BioBERT | Bidirectional Encoder Representations from Transformers for Biomedical Text Mining |
| CORD | COVID-19 Open Research Dataset |
| COVID-19 | COronaVIrus Disease 2019 |
| CPU | central processing unit |
| DB | database |
| DBMS | database management system |
| DOI | digital object identifier |
| FHIR | fast healthcare interoperability resources |
| ID | identifier |
| LBD | literature-based discovery |
| ML | machine learning |
| NER | named entity recognition |
| NLP | natural language processing |
| NoSQL | not only SQL |
| ODQA | open-domain question answering |
| OSI | open systems interconnection |
| PubMed | a search engine for the MEDLINE and some other databases of references and abstracts on life sciences and biomedical topics |
| PubMedBERT | domain-specific language model pretrained for biomedical texts |
| REST | representational state transfer |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome-related Coronavirus 2 |
| SciBERT | Scientific BERT Trained on Semantic Scholar Data |
| SDK | software development kit |
| SeVeN | semantic vector networks |
| SemMedDB | semantic MEDLINE database |
| SemRep | semantic repository |
| SPECTER | scientific paper embeddings using citation-informed transformers |
| SQL | structured query language |
| TBED | text-based emotion detection |
| UID | user identifier |
| UMLS | unified medical language system |
References
- Minaee, S.; Kalchbrenner, N.; Cambria, E.; Nikzad, N.; Chenaghlu, M.; Gao, J. Deep Learning--based Text Classification: A Comprehensive Review. ACM Comput. Surv. 2021, 54, 40. [Google Scholar] [CrossRef]
- Ligthart, A.; Catal, C.; Tekinerdogan, B. Systematic reviews in sentiment analysis: A tertiary study. Artif. Intell. Rev. 2021, 54, 4997–5053. [Google Scholar] [CrossRef]
- Nadeesha, P.; Dehmer, M.; Emmert-Streib, F. Named Entity Recognition and Relation Detection for Biomedical Information Extraction. Front. Cell Dev. Biol. 2020, 8, 673. [Google Scholar] [CrossRef]
- Widyassari, A.P.; Rustad, S.; Shidik, G.F.; Noersasongko, E.; Syukur, A.; Affandy, A. Review of automatic text summarization techniques & methods. J. King Saud Univ.—Comput. Inf. Sci. 2020, 1319–1578. [Google Scholar] [CrossRef]
- Mutabazi, E.; Ni, J.; Tang, G.; Cao, W. A Review on Medical Textual Question Answering Systems Based on Deep Learning Approaches. Appl. Sci. 2021, 11, 5456. [Google Scholar] [CrossRef]
- Zhu, P.; Li, X.; Li, J.; Zhao, H. Unsupervised Open-Domain Question Answering. arXiv 2021, arXiv:2108.13817. [Google Scholar]
- Wang, L.; Lo, K.; Chandrasekhar, Y.; Reas, R.; Yang, J.; Eide, D.; Funk, K.; Kinney, R.; Liu, Z. CORD-19: The Covid-19 Open Research Dataset. arXiv 2020, arXiv:2004.10706v2. [Google Scholar]
- Extance, A. How AI technology can tame the scientific literature. Nature 2018, 561, 273–274. [Google Scholar] [CrossRef] [PubMed]
- Bullock, J.; Luccioni, A.; Pham, K.H.; Lam, C.S.N.; Luengo-Oroz, M. Mapping the landscape of artificial intelligence applications against COVID-19. J. Artif. Inteill. Res. 2020, 69, 807–845. [Google Scholar] [CrossRef]
- Roberts, K.; Alam, T.; Bedrick, S.; Demner-Fushman, D.; Lo, K.; Soboroff, I.; Voorhees, E.; Wang, L.L.; Hersh, W.R. TREC-Covid: Rationale and structure of an information retrieval shared task for covid-19. J. Am. Med. Inform. Assoc. 2020, 27, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Nogueira, R.; Zhang, E.; Gupta, N.; Cam, P.; Cho, K.; Lin, J. Rapidly bootstrapping a question answering dataset for COVID-19. arXiv 2020, arXiv:2004.11339. [Google Scholar]
- Wang, L.L.; Lo, K. Text mining approaches for dealing with the rapidly expanding literature on COVID-19. Brief. Bioinform. 2021, 22, 781–799. [Google Scholar] [CrossRef] [PubMed]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Vol. 1 (Long and Short Papers). Minneapolis, Minnesota: Association for Computational Linguistics; Association for Computational Linguistics: Minneapolis, Minnesota, 2019; pp. 4171–4186. [Google Scholar]
- Beltagy, I.; Lo, K.; Cohan, A. SciBERT: A pretrained language model for scientific text. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP); Association for Computational Linguistics: Hong Kong, China, 2019; pp. 3615–3620. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Yuxian, G.; Robert Tinn, R.; Hao Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. arXiv 2020, arXiv:abs/2007.15779. [Google Scholar]
- National Library of Medicine. Available online: https://pubmed.ncbi.nlm.nih.gov (accessed on 1 December 2021).
- COVID-19 Knowledge Graph. Available online: https://covidgraph.org/ (accessed on 1 December 2021).
- Ilievski, F.; Garijo, D.; Chalupsky, H.; Divvala, N.T.; Yao, Y.; Rogers, C.; Li, R.; Liu, J.; Singh, A.; Schwabe, D.; et al. KGTK: A toolkit for large knowledge graph manipulation and analysis. In Proceedings of the 19th International Semantic Web Conference, Athens, Greece, 2–6 November 2020. [Google Scholar]
- A Free and Open Knowledge Base. Available online: https://www.wikidata.org/ (accessed on 1 December 2021).
- Cohan, A.; Feldman, S.; Beltagy, I.; Downey, D.; Weld, D.S. Specter: Document-level representation learning using citation-informed transformers. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020. [Google Scholar]
- Newman-Griffis, D.; Lai, A.M.; Fosler-Lussier, E. Jointly embedding entities and text with distant supervision. In Proceedings of the Third Workshop on Representation Learning for NLP, Association for Computational Linguistics, Melbourne, Australia, 20 July 2018; pp. 195–206. [Google Scholar]
- Espinosa-Anke, L.; Schockaert, S. SeVeN: Augmenting word embeddings with unsupervised relation vectors. In Proceedings of the 27th International Conference on Computational Linguistics, Santa Fe, NM, USA, 20 August 2018; pp. 2653–2665. [Google Scholar]
- Oniani, D.; Jiang, G.; Liu, H.; Shen, F. Constructing co-occurrence network embeddings to assist association extraction for COVID-19 and other coronavirus infectious diseases. J. Am. Med. Inform. Assoc. 2020, 27, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Unified Medical Language System (UMLS). Available online: https://www.nlm.nih.gov/research/umls/index.html (accessed on 1 December 2021).
- Introducing Text Analytics for Health. Available online: https://techcommunity.microsoft.com/t5/azure-ai/introducing-text-analytics-for-health/ba-p/1505152 (accessed on 1 December 2021).
- Azure Cosmos DB. Available online: https://azure.microsoft.com/en-us/services/cosmos-db/ (accessed on 1 December 2021).
- Python. Available online: https://www.python.org/ (accessed on 1 December 2021).
- The Open Graph Viz Platform. Available online: https://gephi.org (accessed on 1 December 2021).
- Azure Text Analytics Client Library for Python. Available online: https://github.com/Azure/azure-sdk-for-python/blob/main/sdk/textanalytics/azure-ai-textanalytics/README.md (accessed on 1 December 2021).
- Azure Machine Learning. Available online: https://azure.microsoft.com/en-us/services/machine-learning/ (accessed on 1 December 2021).
- COVID-19 Treatment Guidelines. Chloroquine or Hydroxychloroquine and/or Azithromycin. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/chloroquine-or-hydroxychloroquine-and-or-azithromycin/ (accessed on 1 December 2021).
- COVID-19 Treatment Guidelines. Lopinavir/Ritonavir and Other HIV Protease Inhibitors. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/lopinavir-ritonavir-and-other-hiv-protease-inhibitors/ (accessed on 1 December 2021).
- Hamidi Alamdari, D.; Bagheri Moghaddam, A.; Amini, S.; Alamdari, A.H.; Damsaz, M.; Yarahmadi, A. The Application of a Reduced Dye Used in Orthopedics as a Novel Treatment against Coronavirus (COVID-19): A Suggested Therapeutic Protocol. Arch. Bone Jt. Surg. 2020, 8 (Supp. Sl1), 291–294. [Google Scholar] [CrossRef] [PubMed]
- Pundir, H.; Joshi, T.; Joshi, T.; Sharma, P.; Mathpal, S.; Chandra, S.; Tamta, S. Using Chou’s 5-steps rule to study pharmacophore-based virtual screening of SARS-CoV-2 Mpro inhibitors. Mol. Divers. 2021, 25, 1731–1744. [Google Scholar] [CrossRef] [PubMed]
- Caruso, A.; Caccuri, F.; Bugatti, A.; Zani, A.; Vanoni, M.; Bonfanti, P.; Cazzaniga, M.E.; Perno, C.F.; Messa, C.; Alberghina, L. Methotrexate inhibits SARS-CoV-2 virus replication “in vitro”. J. Med. Virol. 2021, 93, 1780–1785. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Treatment Guidelines. Remdesivir. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/remdesivir/ (accessed on 1 December 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).