Usage of the Term Big Data in Biomedical Publications: A Text Mining Approach
Abstract
:1. Introduction
- How well can documents that use the term BD be distinguished from documents that do not use the term in a comparable corpus?
- What are the distinguishing features between BD and NBD documents?
- Does the distinguishability of BD and NBD documents change over time?
2. Related Work
3. Data and Methods
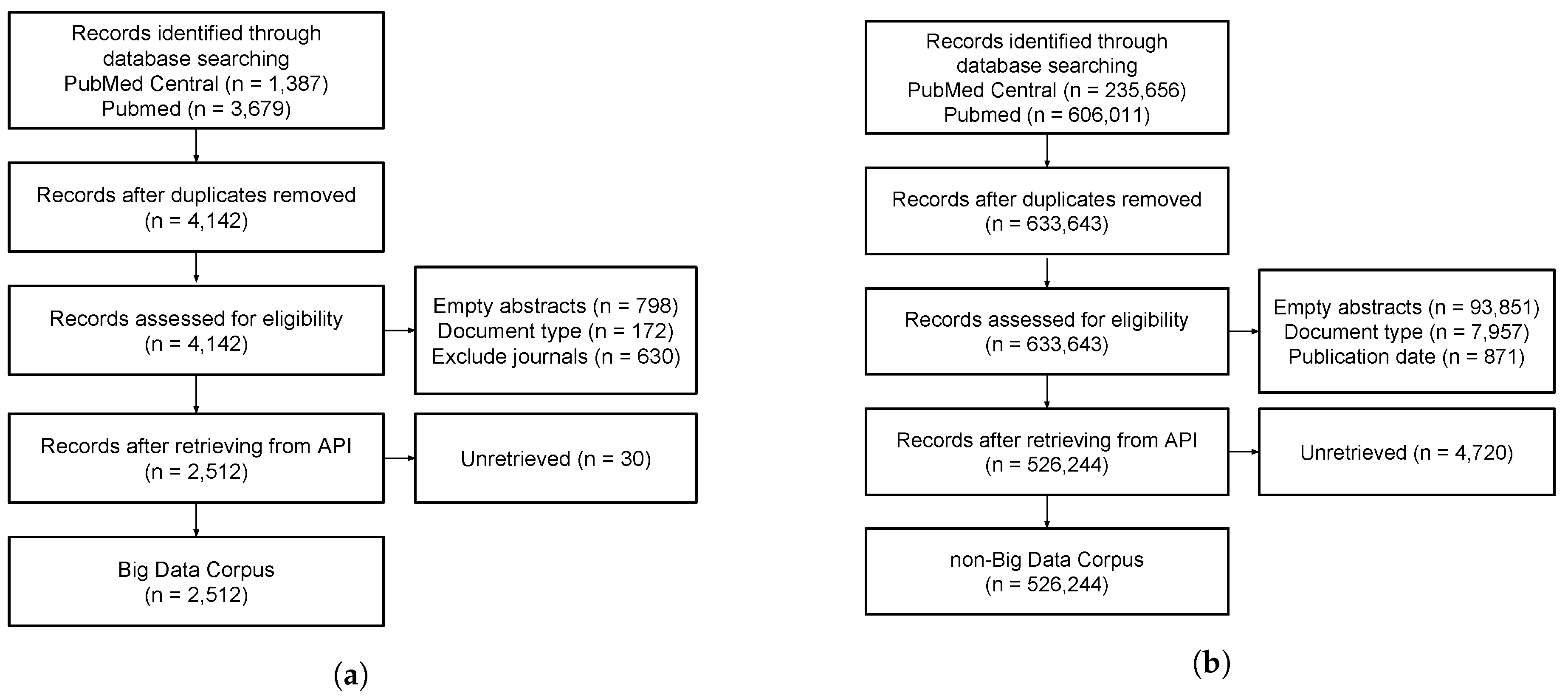
3.1. Corpus Collection
3.1.1. BD Corpus
- PubMed (“Big Data”[TIAB] OR (big[TI] AND data’[TI])).AND (“2011/01/01”[PDAT]: “3000/12/31”[PDAT]) AND English[Language].
- PMC (“Big Data”[TI] OR “Big Data”[AB]) AND (“2011/01/01”[PDAT]: “3000/12/31”[PDAT]).
3.1.2. Non-Big Data Corpus
3.2. Dataset Preparation
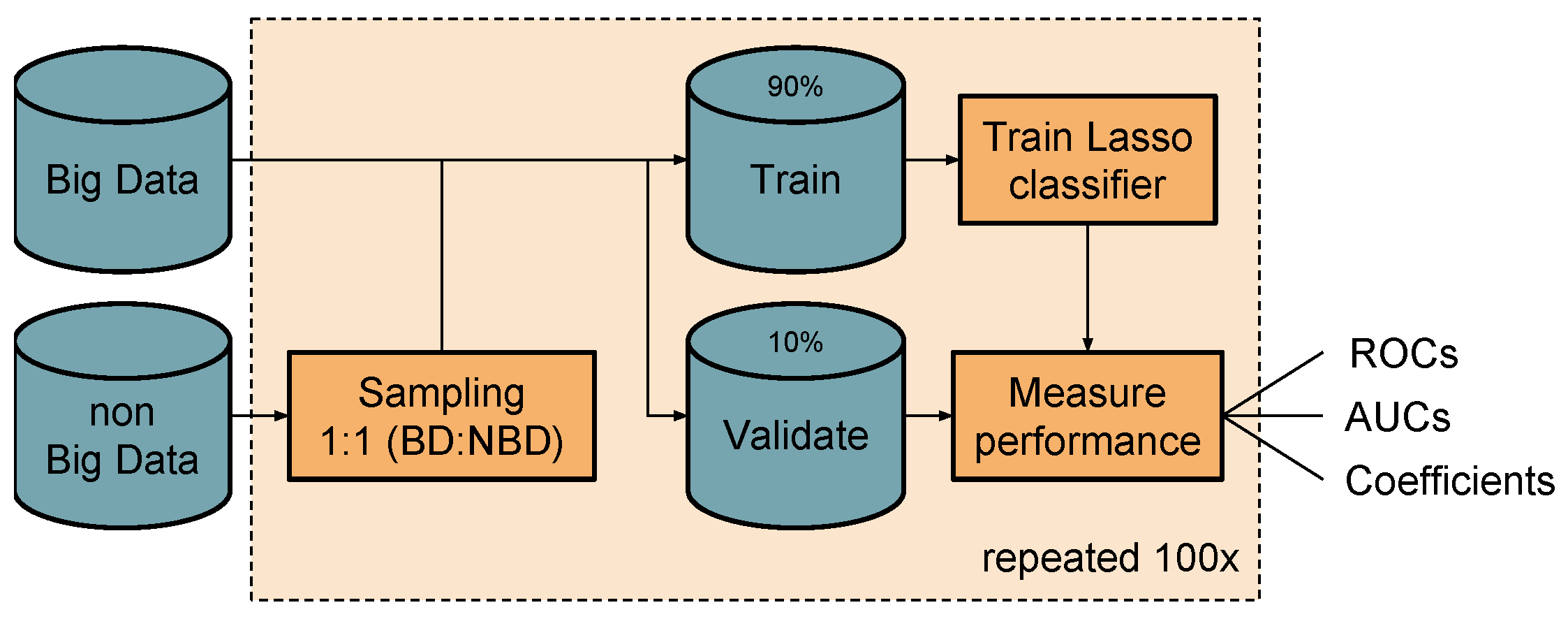
3.3. Classification
Trend Analysis
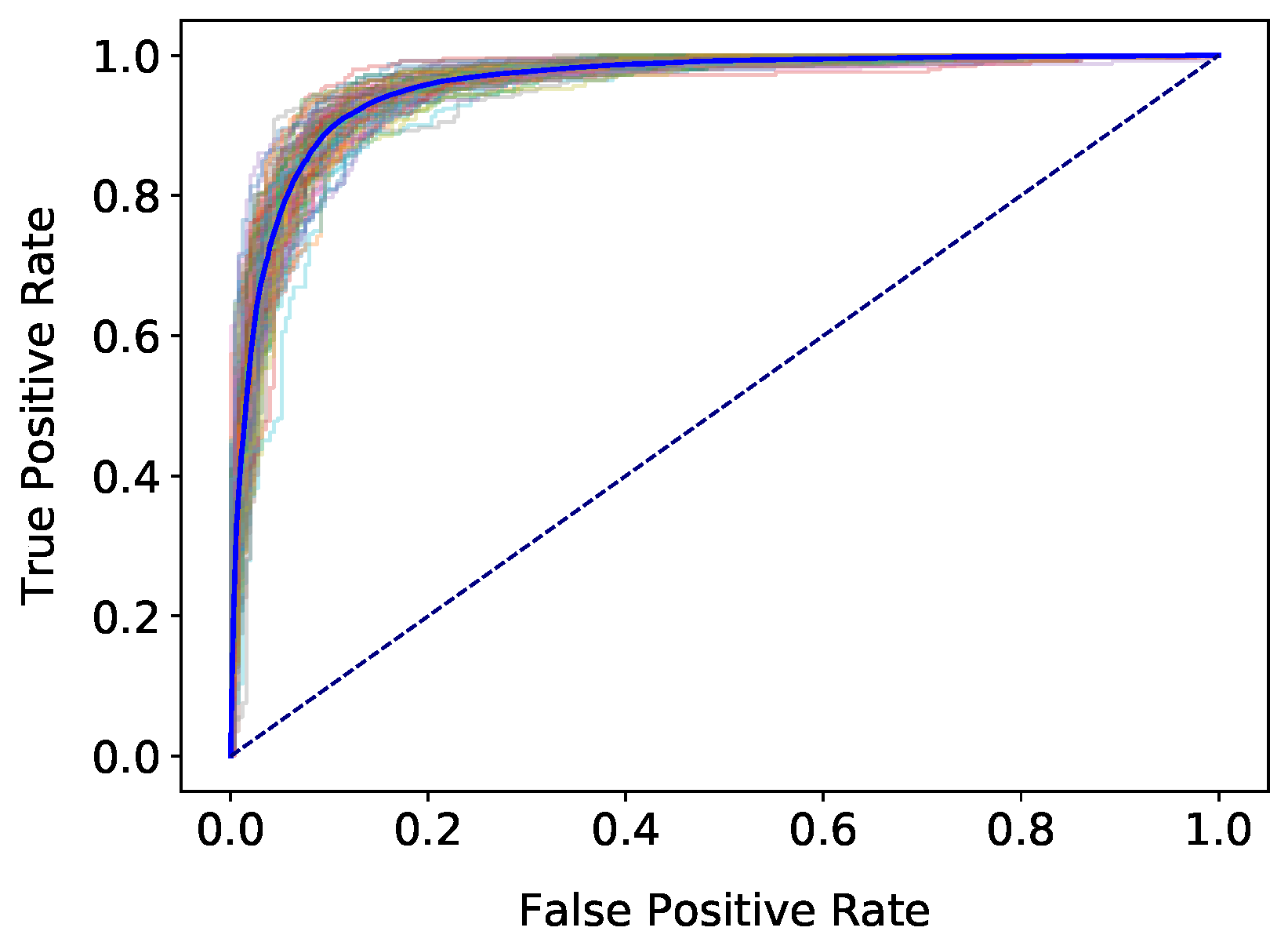
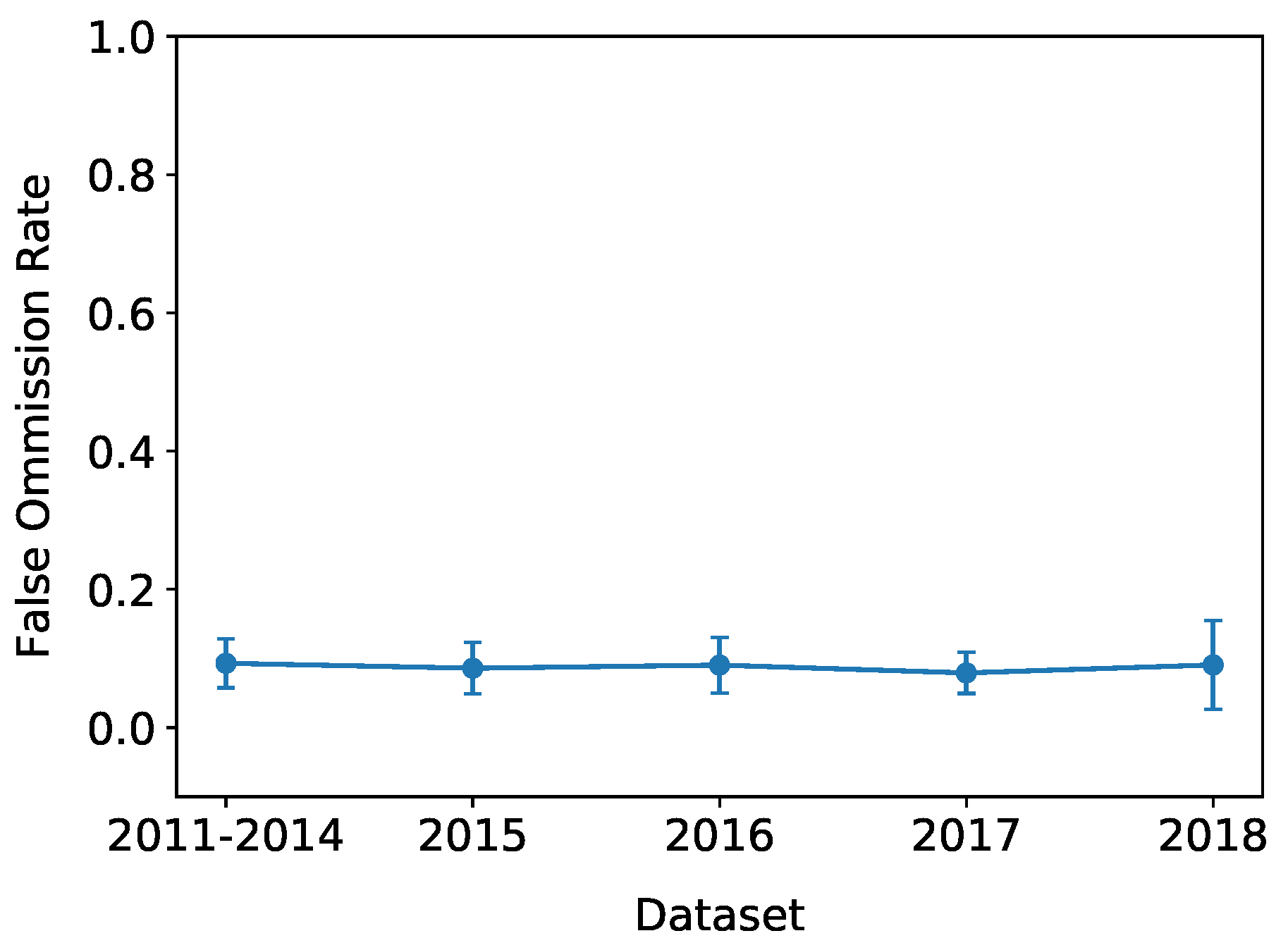
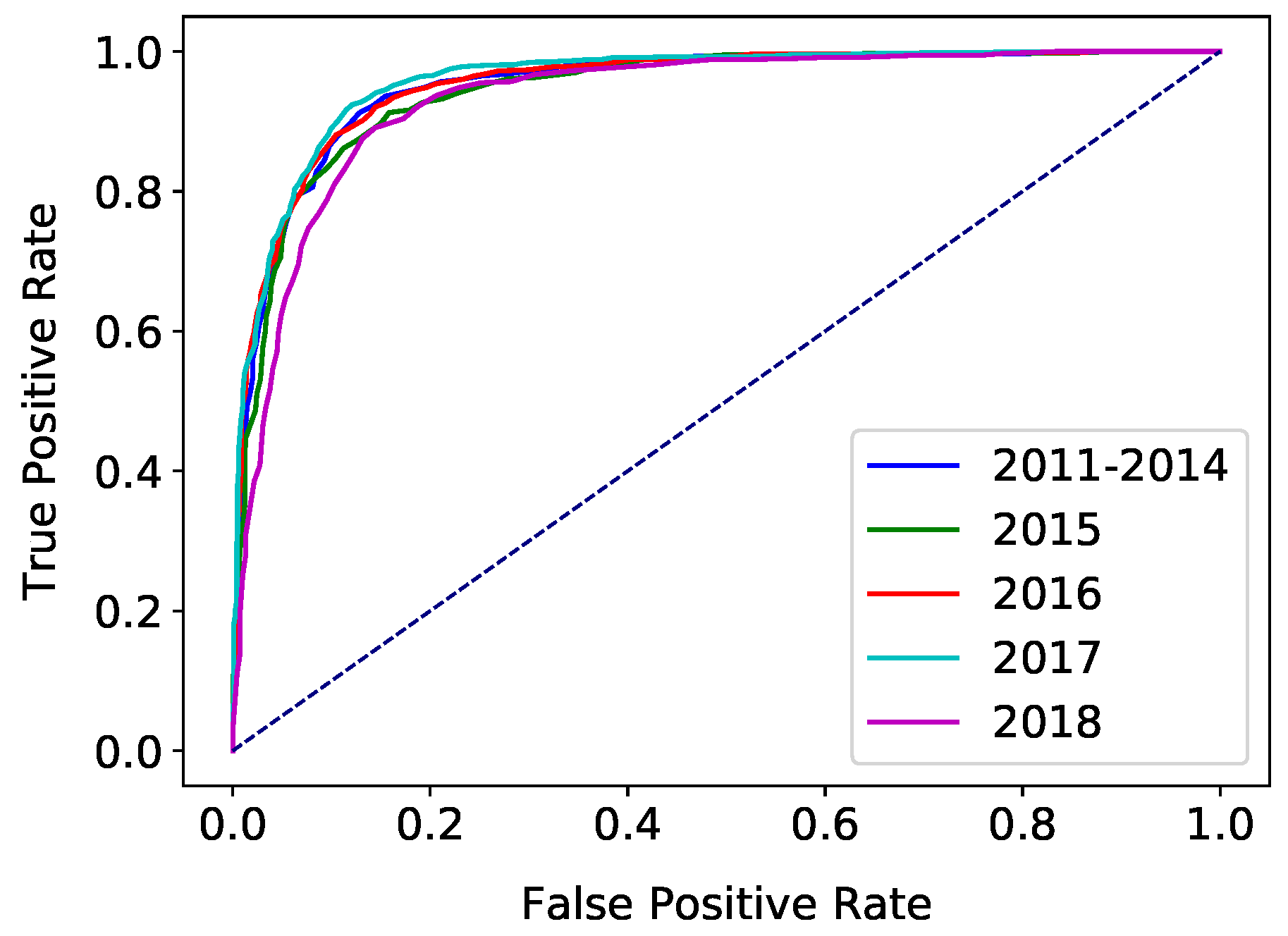
4. Results
5. Discussion
5.1. Corpus, Datasets, and Classification
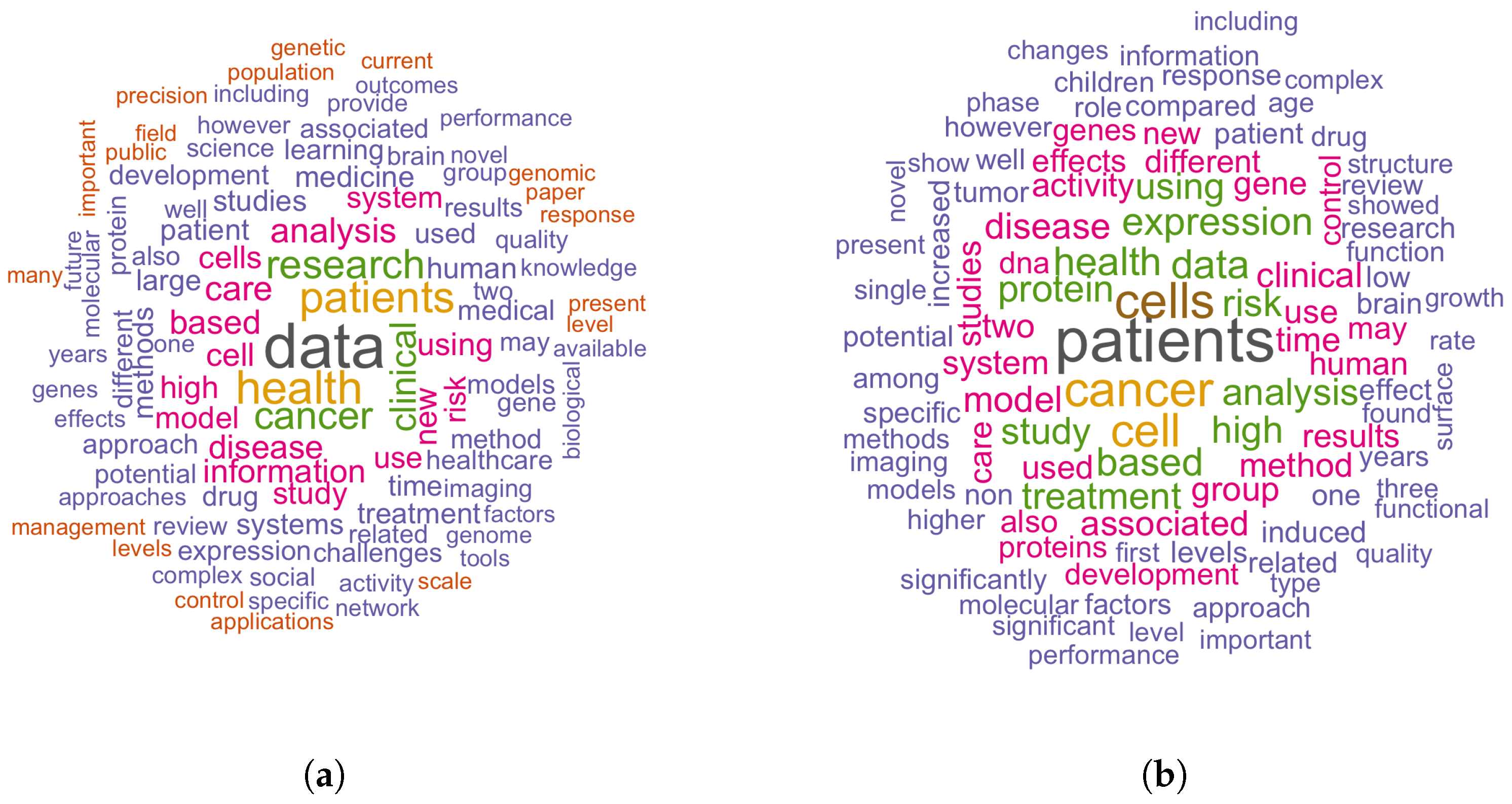
5.2. Distinguishing Features
5.3. Trends Over Time
5.4. Value of the term Big Data
5.5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BD | Big Data |
| NBD | non-Big Data |
| PMC | PubMed Central |
| ROC | Receiver Operating Characteristic |
| AUC | Area Under the Curve |
| FOR | False Omission Rate |
| FDR | False Discovery Rate |
References
- Tian, X. Big data and knowledge management: A case of déjà vu or back to the future? J. Knowl. Manag. 2017, 21, 113–131. [Google Scholar] [CrossRef]
- Mayer-Schönberger, V.; Cukier, K. Big Data: A Revolution that Will Transform how We Live, Work, and Think; Mariner Books: Wilmington, DE, USA, 2013; p. 257. [Google Scholar]
- Hahn, A.; Mohanty, S.D.; Manda, P. What’s Hot and What’s Not? - Exploring Trends in Bioinformatics Literature Using Topic Modeling and Keyword Analysis. In Bioinformatics Research and Applications, Proceedings of the 13th International Symposium, ISBRA 2017, Honolulu, HI, USA, 29 May–2 June 2017, Proceedings; Cai, Z., Daescu, O., Li, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 279–290. [Google Scholar] [CrossRef]
- Weiss, S.; Indurkhya, N.; Zhang, T.; J. Damerau, F. Text Mining: Predictive Methods for Analyzing Unstructured Information; Springer Science & Business Media: Berlin, Germany, 2004. [Google Scholar]
- Zhou, H.k.; Yu, H.m.; Hu, R. Topic discovery and evolution in scientific literature based on content and citations. Front. Inf. Technol. Electron. Eng. 2017, 18, 1511–1524. [Google Scholar] [CrossRef]
- van Altena, A.J.; Moerland, P.D.; Zwinderman, A.H.; Olabarriaga, S.D. Analysis of the term ‘big data’: Usage in biomedical publications. In Proceedings of the 2017 IEEE International Conference on Big Data (Big Data), Boston, MA, USA, 11–14 December 2017; pp. 1253–1258. [Google Scholar] [CrossRef]
- Laney, D. 3D data management: Controlling data volume, velocity and variety. META Group Res. Note 2001, 6, 70. [Google Scholar]
- Andreu-Perez, J.; Poon, C.C.; Merrifield, R.D.; Wong, S.T.; Yang, G.Z. Big Data for Health. IEEE J. Biomed. Health Inform. 2015, 19, 1193–1208. [Google Scholar] [CrossRef] [PubMed]
- Levi, M. Kleren van de Keizer [The Emperor’s Clothes]. Column, Medisch Contact. 2015. Available online: https://www.medischcontact.nl/opinie/blogs-columns/column/kleren-van-de-keizer-marcel-levi.htm (accessed on 12 March 2018).
- Heudecker, N. Big Data Isn’t Obsolete. It’s Normal. 2015. Available online: http://blogs.gartner.com/nick-heudecker/big-data-is-now-normal/ (accessed on 12 March 2018).
- Foo, A. Face It, Big Data Is the New Normal. 2013. Available online: http://www.ibmbigdatahub.com/blog/face-it-big-data-new-normal (accessed on 12 March 2018).
- Anon. Big Data Series. 2014. Available online: https://www.parliament.uk/mps-lords-and-offices/offices/bicameral/post/work-programme/big-data/ (accessed on 12 March 2018).
- Laney, D. Big Data’s 10 Biggest Vision and Strategy Questions. 2015. Available online: http://blogs.gartner.com/doug-laney/big-datas-10-biggest-vision-and-strategy-questions/ (accessed on 12 March 2018).
- De Mauro, A.; Greco, M.; Grimaldi, M. A formal definition of Big Data based on its essential features. Libr. Rev. 2016, 65, 122–135. [Google Scholar] [CrossRef]
- Ward, J.S.; Barker, A. Undefined By Data: A Survey of Big Data Definitions. arXiv, 2013; arXiv:1309.5821. [Google Scholar]
- Gandomi, A.; Haider, M. Beyond the hype: Big data concepts, methods, and analytics. Int. J. Inf. Manag. 2015, 35, 137–144. [Google Scholar] [CrossRef]
- Kudva, S.; Ye, X. Smart Cities, Big Data, and Sustainability Union. Big Data Cognit. Comput. 2017, 1. [Google Scholar] [CrossRef]
- Wolfert, S.; Ge, L.; Verdouw, C.; Bogaardt, M.J. Big Data in Smart Farming—A review. Agric. Syst. 2017, 153, 69–80. [Google Scholar] [CrossRef]
- Hashem, I.A.T.; Yaqoob, I.; Anuar, N.B.; Mokhtar, S.; Gani, A.; Khan, S.U. The rise of “big data” on cloud computing: Review and open research issues. Inf. Syst. 2015, 47, 98–115. [Google Scholar] [CrossRef]
- Hansmann, T.; Niemeyer, P. Big Data—Characterizing an Emerging Research Field Using Topic Models. In Proceedings of the 2014 IEEE/WIC/ACM International Joint Conferences on Web Intelligence (WI) and Intelligent Agent Technologies (IAT), Warsaw, Poland, 11–14 August 2014; IEEE Computer Society: Washington, DC, USA, 2014; Volume 1, pp. 43–51. [Google Scholar]
- van Altena, A.J.; Moerland, P.D.; Zwinderman, A.H.; Olabarriaga, S.D. Understanding big data themes from scientific biomedical literature through topic modeling. J. Big Data 2016, 3, 23. [Google Scholar] [CrossRef]
- Kitchin, R. Big data and human geography: Opportunities, challenges and risks. Dialogues Hum. Geogr. 2013, 3, 262–267. [Google Scholar] [CrossRef]
- Kitchin, R.; McArdle, G. What makes Big Data, Big Data? Exploring the ontological characteristics of 26 datasets. Big Data Soc. 2016, 3, 2053951716631130. [Google Scholar] [CrossRef]
- van Altena, A.J. AMCeScience/python-miner-pub. 2019. Available online: https://github.com/AMCeScience/python-miner-pub/ (accessed on 4 February 2019).
- Bethesda (MD): National Center for Biotechnology Information (US). Entrez Programming Utilities Help. 2010. Available online: https://www.ncbi.nlm.nih.gov/books/NBK25501/ (accessed on 18 May 2018).
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. J. Clin. Epidemiol. 2009, 62, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Loper, E.; Bird, S. NLTK: The Natural Language Toolkit. In Proceedings of the ACL Workshop on Effective Tools and Methodologies for Teaching Natural Language Processing and Computational Linguistics, Philadelphia, PA, USA, 7 July 2002; Association for Computational Linguistics: Philadelphia, PA, USA, 2002. [Google Scholar]
- Van Altena, A.J. AMCeScience/R-contrast-pub. 2019. Available online: https://github.com/AMCeScience/R-contrast-pub/ (accessed on 4 February 2019).
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]

| BD | non-Big Data | |
|---|---|---|
| # docs | 2512 | 526,244 |
| # journals | 1189 | 1144 |
| # docs per journal * | 2 [1–73] | 460 [1–10,298] |
| # docs per year | ||
| 2011 | 5 | 839 |
| 2012 | 18 | 5668 |
| 2013 | 100 | 23,825 |
| 2014 | 271 | 53,307 |
| 2015 | 411 | 87,220 |
| 2016 | 631 | 134,876 |
| 2017 | 728 | 175,590 |
| 2018 | 348 | 44,919 |
| # tokens | ||
| all * | 133 [13–516] | 139 [0–1210] |
| title * | 9 [1–28] | 10 [0–57] |
| abstract * | 125 [10–511] | 129 [0–1205] |
| # unique tokens | ||
| all * | 94 [12–287] | 91 [0–425] |
| title * | 9 [1–24] | 10 [0–48] |
| abstract * | 92 [10–287] | 89 [0–424] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Altena, A.J.; Moerland, P.D.; Zwinderman, A.H.; Delgado Olabarriaga, S. Usage of the Term Big Data in Biomedical Publications: A Text Mining Approach. Big Data Cogn. Comput. 2019, 3, 13. https://doi.org/10.3390/bdcc3010013
van Altena AJ, Moerland PD, Zwinderman AH, Delgado Olabarriaga S. Usage of the Term Big Data in Biomedical Publications: A Text Mining Approach. Big Data and Cognitive Computing. 2019; 3(1):13. https://doi.org/10.3390/bdcc3010013
Chicago/Turabian Stylevan Altena, Allard J., Perry D. Moerland, Aeilko H. Zwinderman, and Sílvia Delgado Olabarriaga. 2019. "Usage of the Term Big Data in Biomedical Publications: A Text Mining Approach" Big Data and Cognitive Computing 3, no. 1: 13. https://doi.org/10.3390/bdcc3010013
APA Stylevan Altena, A. J., Moerland, P. D., Zwinderman, A. H., & Delgado Olabarriaga, S. (2019). Usage of the Term Big Data in Biomedical Publications: A Text Mining Approach. Big Data and Cognitive Computing, 3(1), 13. https://doi.org/10.3390/bdcc3010013





