Multidrug-Resistant Proteus mirabilis and Other Gram-Negative Species Isolated from Native Egyptian Chicken Carcasses
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Preparation of Samples
2.2. Isolation of Presumptive Typical Salmonella Colonies on XLD
2.3. Molecular Testing Conducted to Allow Differentiation between Salmonella and Other Gram-Negative Competitor Bacteria
2.4. Biochemical Identification
2.5. Serological Identification
2.6. Antibiotic Susceptibility Testing for the Identified Isolates
3. Results and Discussion
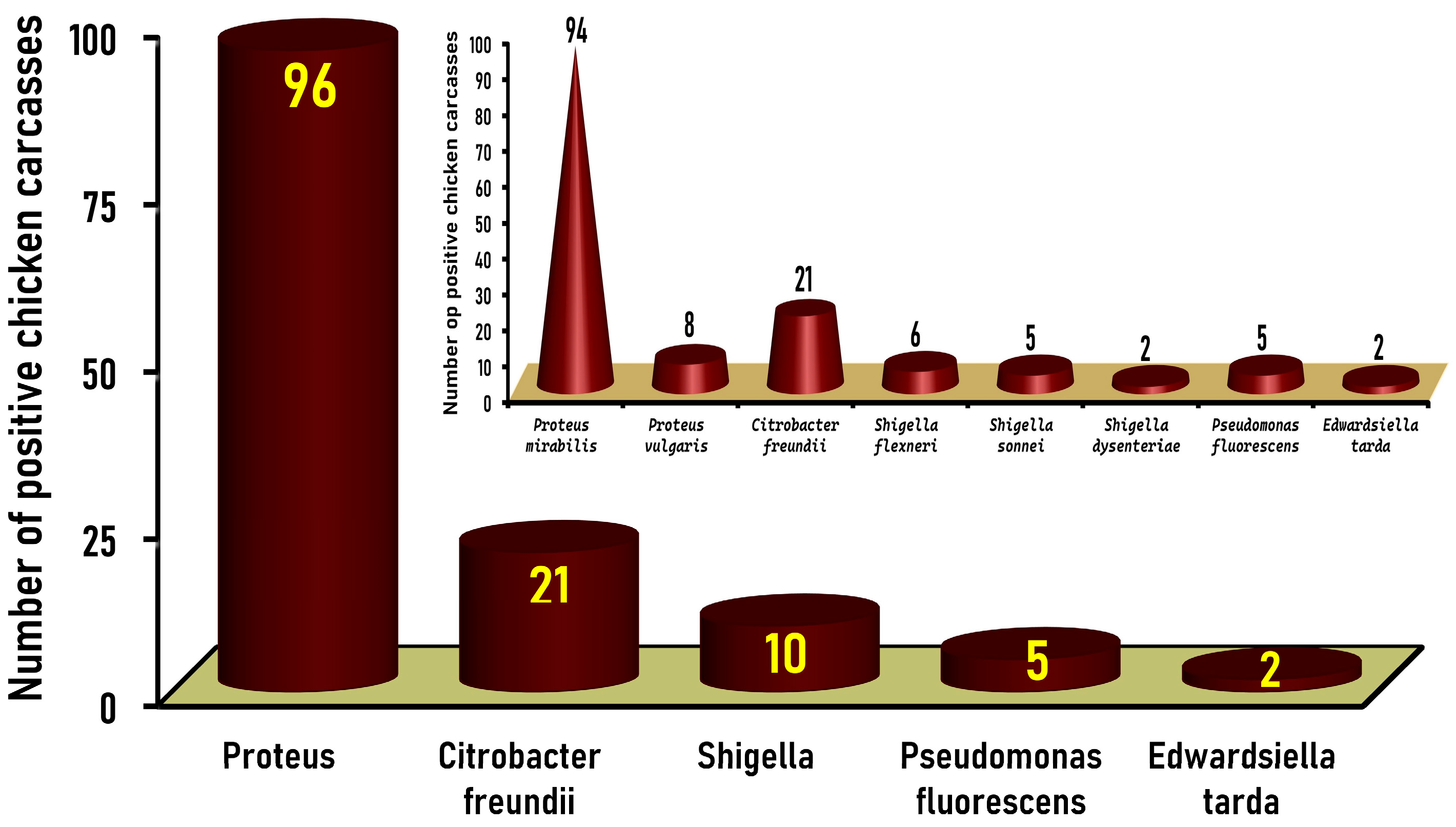
3.1. Prevalence of Gram-Negative Non-Salmonella Isolates in Native Egyptian Chicken Carcasses
3.2. Frequency Distribution of the Identified Gram-Negative Non-Salmonella Isolates Recovered from Native Egyptian Chicken Carcasses
3.3. Antimicrobial Resistance of the Identified Species of Gram-Negative, Non-Salmonella Isolates Recovered from Native Egyptian Chicken Carcasses
3.4. Categorization of the Different Identified Gram-Negative (Non-Salmonella) Species Based on Their Antibiotic Resistance Profiles and Their Multiple Antibiotic Resistance (MAR) Index Values
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parija, S.C. Textbook of Microbiology and Immunology; Springer Nature: Singapore, 2023. [Google Scholar] [CrossRef]
- Galal, S. Annual Chicken Meat Production in Egypt 2010–2023. Available online: https://www.statista.com/statistics/1005988/egypt-chicken-meat-production/#:~:text=In%202023%2C%20chicken%20meat%20production,in%20the%20period%20under%20review (accessed on 6 September 2024).
- Sun, T.; Liu, Y.; Qin, X.; Aspridou, Z.; Zheng, J.; Wang, X.; Li, Z.; Dong, Q. The prevalence and epidemiology of Salmonella in retail raw poultry meat in China: A systematic review and meta-analysis. Foods 2021, 10, 2757. [Google Scholar] [CrossRef] [PubMed]
- Adzitey, F.; Huda, N.; Gulam, R. Comparison of media for the isolation of Salmonella (XLD and Rambach) and Listeria species (ALOA and Palcam) in naturally contaminated duck samples. Internet J. Food Saf. 2011, 13, 20–25. [Google Scholar]
- Sun, Y.; Wen, S.; Zhao, L.; Xia, Q.; Pan, Y.; Liu, H.; Wei, C.; Chen, H.; Ge, J.; Wang, H. Association among Biofilm Formation, virulence gene expression, and antibiotic resistance in Proteus mirabilis isolates from diarrhetic animals in Northeast China. BMC Vet. Res. 2020, 16, 176. [Google Scholar] [CrossRef]
- Schaffer, J.N.; Pearson, M.M. Proteus mirabilis and urinary tract infections. Microbiol. Spectr. 2015, 3, 383–433. [Google Scholar] [CrossRef]
- Van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef]
- Zaidi, M.B.; Estrada-García, T. Shigella: A highly virulent and elusive pathogen. Curr. Trop. Med. Rep. 2014, 1, 81–87. [Google Scholar] [CrossRef]
- Park, Y.-J.; Yu, J.K.; Lee, S.; Oh, E.-J.; Woo, G.-J. Prevalence and diversity of qnr alleles in Ampc-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: A multicentre study from Korea. J. Antimicrob. Chemother. 2007, 60, 868–871. [Google Scholar] [CrossRef]
- Samonis, G.; Karageorgopoulos, D.E.; Kofteridis, D.P.; Matthaiou, D.K.; Sidiropoulou, V.; Maraki, S.; Falagas, M.E. Citrobacter infections in a general hospital: Characteristics and outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lan, R.; Liu, L.; Wang, Y.; Zhang, Y.; Wang, Y.; Xu, J. Antimicrobial resistance and cytotoxicity of Citrobacter spp. in Maanshan Anhui Province, China. Front. Microbiol. 2017, 8, 1357. [Google Scholar] [CrossRef]
- An, L.; Chan, J.L.; Nguyen, M.; Yang, S.; Deville, J.G. Case report: Disseminated Edwardsiella tarda infection in an immunocompromised patient. Front. Cell Infect. Microbiol. 2023, 13, 1292768. [Google Scholar] [CrossRef]
- Gershman, M.D.; Kennedy, D.J.; Noble-Wang, J.; Kim, C.; Gullion, J.; Kacica, M.; Jensen, B.; Pascoe, N.; Saiman, L.; McHale, J.; et al. Multistate outbreak of Pseudomonas fluorescens bloodstream infection after exposure to contaminated heparinized saline flush prepared by a compounding pharmacy. Clin. Infect. Dis. 2008, 47, 1372–1379. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.J.; Ray, K.G. Sherris Medical Microbiology, 5th ed.; McGraw-Hill Medical: New York, NY, USA, 2010. [Google Scholar]
- Hedman, H.D.; Vasco, K.A.; Zhang, L. A review of antimicrobial resistance in poultry farming within low-resource settings. Animals 2020, 10, 1264. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Njoga, E.O.; Nwanta, J.A.; Chah, K.F. Detection of multidrug-resistant Campylobacter species from food-producing animals and humans in Nigeria: Public health implications and one health control measures. Comp. Immunol. Microbiol. Infect. Dis. 2023, 103, 102083. [Google Scholar] [CrossRef]
- USDA/FSIS Isolation and Identification of Salmonella from Meat, Poultry, Pasteurized Egg, Carcass, and Environmental Sponges. United States Department of Agriculture Food Safety and Inspection Service MLG 4.13. 2023. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/documents/MLG-4.13.pdf (accessed on 18 October 2023).
- Chiu, C.H.; Ou, J.T. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 1996, 34, 2619–2622. [Google Scholar] [CrossRef]
- Carroll, K.C.; Pfaller, M.A.; Karlowsky, J.A.; Landry, M.L.; Mcadam, A.J.; Patel, R.; Pritt, B.S. (Eds.) Manual of Clinical Microbiology, 13th ed.; ASM Press: Washington, DC, USA, 2019. [Google Scholar]
- Knirel, Y.A.; Perepelov, A.V.; Kondakova, A.N.; Senchenkova, S.N.; Sidorczyk, Z.; Rozalski, A.; Kaca, W. Structure and serology of O-antigens as the basis for classification of Proteus strains. Innate Immun. 2011, 17, 70–96. [Google Scholar] [CrossRef]
- Lányi, B. Lányi, B. 2 Biochemical and serological characterization of Citrobacter. In Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 15, pp. 143–171. [Google Scholar]
- Abbott, S.L. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other Enterobacteriaceae. In Manual of Clinical Microbiology, 10th ed.; Versalovic, J., Carroll, K.C., Funke, G., Jorgensen, J.H., Landry, M.L., Warnock, D.W., Eds.; Ed. American Society of Microbiology: Washington, DC, USA, 2011; Chapter 37; pp. 639–657. [Google Scholar]
- Pitt, T.L.; Erdman, Y.J. The specificity of agglutination reactions of Pseudomonas aeruginosa with O antisera. J. Med. Microbiol. 1978, 11, 15–23. [Google Scholar] [CrossRef]
- Ansorg, R.; Knoche, M. Determination of the O-serovars of Pseudomonas aeruginosa by slide coagglutination. Eur. J. Clin. Microbiol. 1984, 3, 190–194. [Google Scholar] [CrossRef]
- CLSI “Clinical and Laboratory Standards Institute”. Performance Standards for Antimicrobial Susceptibility Testing, M100, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Singh, S.; Yadav, A.S.; Singh, S.M.; Bharti, P. Prevalence of Salmonella in chicken eggs collected from poultry farms and marketing channels and their antimicrobial resistance. Food Res. Int. 2010, 43, 2027–2030. [Google Scholar] [CrossRef]
- El-Saeed, B.A.; Elshebrawy, H.A.; Zakaria, A.I.; Abdelkhalek, A.; Sallam, K.I. Colistin-, cefepime-, and levofloxacin-resistant Salmonella enterica serovars isolated from Egyptian chicken carcasses. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 61. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.W.; Zhang, A.Y.; Wang, H.N.; Liu, B.H.; Yang, L.Q.; Yang, Y.Q. Characterization of SXT/R391 integrative and conjugative elements in Proteus mirabilis isolates from food-producing animals in China. Antimicrob. Agents Chemother. 2016, 60, 1935–1938. [Google Scholar] [CrossRef] [PubMed]
- Firildak, G.; Asan, A.; Goren, E. Chicken carcasses bacterial concentration at poultry slaughtering facilities. Asian J. Biol. Sci. 2015, 8, 16–29. [Google Scholar] [CrossRef]
- Wong, M.H.Y.; Wan, H.Y.; Chen, S. Characterization of multidrug-resistant Proteus mirabilis isolated from chicken carcasses. Foodborne Pathog. Dis. 2013, 10, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Yulistiani, R.; Praseptiangga, D. Contamination level and prevalence of foodborne pathogen Enterobacteriaceae in broiler and backyard chicken meats sold at traditional markets in Surabaya, Indonesia. Malays. Appl. Biol. 2019, 48, 95–103. [Google Scholar]
- Yu, Z.; Joossens, M.; Van Den Abeele, A.-M.; Kerkhof, P.-J.; Houf, K. Isolation, Characterization and antibiotic resistance of Proteus mirabilis from Belgian broiler carcasses at retail and human stool. Food Microbiol. 2021, 96, 103724. [Google Scholar] [CrossRef]
- Ishaq, K. Occurrence and antimicrobial susceptibility of Proteus mirabilis from chicken carcass. PVJ 2022, 42, 576–579. [Google Scholar] [CrossRef]
- Barbour, E.K.; Hajj, Z.G.; Hamadeh, S.; Shaib, H.A.; Farran, M.T.; Araj, G.; Faroon, O.; Barbour, K.E.; Jirjis, F.; Azhar, E.; et al. Comparison of phenotypic and virulence genes characteristics in human and chicken isolates of Proteus mirabilis. Pathog. Glob. Health. 2012, 106, 352–357. [Google Scholar] [CrossRef]
- Afify, S.; Shaltout, F.; Mohammed, I. Bacteriological profile of some raw chicken meat cuts in Ismailia City, Egypt. Benha Vet. Med. J. 2020, 39, 11–15. [Google Scholar] [CrossRef]
- Noori, T.E.; Alwan, M.J. Isolation and identification of zoonotic bacteria from poultry meat. Int. J. Adv. Res. Biol. Sci. 2016, 3, 57–66. [Google Scholar]
- Saud, B.; Paudel, G.; Khichaju, S.; Bajracharya, D.; Dhungana, G.; Awasthi, M.S.; Shrestha, V. Multidrug-resistant bacteria from raw meat of buffalo and chicken, Nepal. Vet. Med. Int. 2019, 2019, 7960268. [Google Scholar] [CrossRef]
- Qader, M.B.A.; AlKhafaji, M.H. Detection of bacterial contamination of imported chicken meat in Iraq. Iraqi J. Sci. 2019, 60, 1957–1966. [Google Scholar] [CrossRef]
- Al-Asbahi, A.A. Prevalence and Bacteriological Study of Gram-negative bacteria especially Citrobacter spp. in Poultry meat from Maeen Area- Sana’a, Yemen. J. Appl. Vet. Sci. 2022, 7, 62–66. [Google Scholar]
- Hashim, M.H.; AlKhafaji, M.H. Isolation and identification of Citrobacter freundii from chicken meat samples using cultural and molecular techniques. Iraqi J. Sci. 2018, 59, 1216–1224. [Google Scholar] [CrossRef]
- Sánchez-Códez, M.I.; Alonso-Ojembarrena, A.; Arca-Suárez, J. Gramnegativos infrecuentes como agentes etiológicos de infecciones nosocomiales en una Unidad de Cuidados Intensivos Neonatales. Rev. Esp. Quim. 2018, 31, 288–290. [Google Scholar]
- Ahmed, A.M.; Shimamoto, T. Isolation and molecular characterization of Salmonella enterica, Escherichia coli O157: H7 and Shigella spp. from meat and dairy products in Egypt. Int. J. Food Microbiol. 2014, 168, 57–62. [Google Scholar] [CrossRef]
- Sackey, B.A.; Mensah, P.; Collison, E.; Sakyi-Dawson, E. Campylobacter, Salmonella, Shigella and Escherichia coli in live and dressed poultry from metropolitan accra. Int. J. Food Microbiol. 2001, 71, 21–28. [Google Scholar] [CrossRef]
- Tagar, S.; Qambrani, N.A. Bacteriological quality assessment of poultry chicken meat and meat contact surfaces for the presence of targeted bacteria and determination of antibiotic resistance of Salmonella spp. in Pakistan. Food Control 2023, 151, 109786. [Google Scholar] [CrossRef]
- Rabins, L. Epidemiological study on Shigella from meat and its public health significance. Int. J. Curr. Microbiol. App. Sci. 2021, 10, 433–443. [Google Scholar] [CrossRef]
- Cetinkaya, F.; Cibik, R.; Ece Soyutemiz, G.; Ozakin, C.; Kayali, R.; Levent, B. Shigella and Salmonella contamination in various foodstuffs in Turkey. Food Control 2008, 19, 1059–1063. [Google Scholar] [CrossRef]
- WHO “World Health Organization”. Shigellosis: Disease burden, epidemiology and case management. Relev. Epidemiol. Hebd. 2005, 80, 94–99. [Google Scholar]
- Heir, E.; Moen, B.; Åsli, A.W.; Sunde, M.; Langsrud, S. Antibiotic resistance and phylogeny of Pseudomonas spp. isolated over three decades from chicken meat in the Norwegian food chain. Microorganisms 2021, 9, 207. [Google Scholar] [CrossRef]
- El-Aziz, A. Detection of Pseudomonas spp. in chicken and fish sold in markets of Assiut City, Egypt. J. Food Qual. Hazards Control 2015, 2, 86–89. [Google Scholar]
- Can, H.Y. Investigation of Pseudomonas species in chicken drumstick samples. Kocatepe Vet. J. 2022, 15, 139–143. [Google Scholar] [CrossRef]
- Elbehiry, A.; Marzouk, E.; Aldubaib, M.; Moussa, I.; Abalkhail, A.; Ibrahem, M.; Hamada, M.; Sindi, W.; Alzaben, F.; Almuzaini, A.M.; et al. Pseudomonas species prevalence, protein analysis, and antibiotic resistance: An evolving public health challenge. AMB Expr. 2022, 12, 53. [Google Scholar] [CrossRef]
- Samir, S.; Awad, A.; Younis, G. Prevalence, virulence determinants and antimicrobial-resistant Profile of Edwardsiella tarda isolated from Nile tilapia (Oreochromis niloticus) in Egypt. AAVS 2021, 10, 1031–1038. [Google Scholar] [CrossRef]
- Shrestha, A.; Bajracharya, A.M.; Subedi, H.; Turha, R.S.; Kafle, S.; Sharma, S.; Neupane, S.; Chaudhary, D.K. Multi-drug resistance and extended spectrum beta lactamase producing Gram negative bacteria from chicken meat in Bharatpur Metropolitan, Nepal. BMC Res. Notes 2017, 10, 574. [Google Scholar] [CrossRef]
- Mberu, C. Salmonella and Shigella Species Associated with Broiler Chicken Meat and Their Susceptibility Patterns to Antimicrobials. Repository.mouau.edu.ng: 2021. Available online: https://repository.mouau.edu.ng/work/view/salmonella-and-shigella-species-associated-with-broiler-chicken-meat-and-their-susceptbility-patterns-to-antimicrobials-7-2 (accessed on 11 January 2024).
- Odoi, J.O.; Takayanagi, S.; Sugiyama, M.; Usui, M.; Tamura, Y.; Asai, T. prevalence of colistin-resistant bacteria among retail meats in Japan. Food Saf. 2021, 9, 48–56. [Google Scholar] [CrossRef]
- Abd El-Tawab, A.A.; Selim, A.O.; Soliman, A.M. Phenotypic and genotypic characterization of some bacterial isolates (Escherichia coli, Klebsiella oxytoca) from chickens. BVMJ 2018, 35, 284–302. [Google Scholar] [CrossRef]
- Wang, X.; Yan, M.; Wang, Q.; Ding, L.; Li, F. Identification of Edwardsiella tarda isolated from duck and virulence genes detection. Afr. J. Microbiol. Res. 2012, 6, 4970–4975. [Google Scholar] [CrossRef]
- CDC “Centers for Disease Control and Prevention”. Gram-Negative Bacteria Infections in Healthcare Settings; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2011.
- Yulistiani, R.; Praseptiangga, D.; Supyani; Sudibya; Raharjo, D.; Shirakawa, T. Prevalence of antibiotic-resistance Enterobacteriaceae strains isolated from chicken meat at traditional markets in Surabaya, Indonesia. IOP Conf. Ser. Mater. Sci. Eng. 2017, 193, 012007. [Google Scholar] [CrossRef]
- Li, Z.; Peng, C.; Zhang, G.; Shen, Y.; Zhang, Y.; Liu, C.; Liu, M.; Wang, F. Prevalence and characteristics of multidrug-resistant Proteus mirabilis from broiler farms in Shandong Province, China. Poult. Sci. 2022, 101, 101710. [Google Scholar] [CrossRef] [PubMed]
- Moktan, J.B.; Venkataraman, R.; Shrestha, Y. The prevalence of multidrug-resistant bacteria detected in poultry products in Mandya, India. Arch. Pharm. Pract. 2023, 14, 35–39. [Google Scholar] [CrossRef]


| Proteus (n = 143) | Citrobacter (n = 26) | Shigella (n = 17) | Pseudomonas (n = 6) | Edwardsiella (n = 3) | Total (n = 195) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| Cefaclor (CEC) | 143 | 100 | 26 | 100 | 17 | 100 | 6 | 100 | 3 | 100 | 195 | 100 |
| Fosfomycin (FOS) | 143 | 100 | 26 | 100 | 17 | 100 | 6 | 100 | 3 | 100 | 195 | 100 |
| Cephalothin (KF) | 143 | 100 | 22 | 84.6 | 17 | 100 | 6 | 100 | 3 | 100 | 191 | 98 |
| Azithromycin (AZM) | 134 | 93.7 | 26 | 100 | 12 | 70.6 | 6 | 100 | 3 | 100 | 181 | 92.8 |
| Vancomycin (VA) | 128 | 89.5 | 26 | 100 | 12 | 70.6 | 6 | 100 | 3 | 100 | 175 | 89.7 |
| Nalidixic acid (NA) | 133 | 93 | 21 | 80.8 | 15 | 88.2 | 4 | 66.7 | 1 | 33.3 | 174 | 89.2 |
| Tetracycline (TE) | 132 | 92.3 | 21 | 80.8 | 12 | 70.6 | 6 | 100 | 3 | 100 | 174 | 89.2 |
| Sulfamethoxazole/Trimethoprim (SXT) | 127 | 88.8 | 21 | 80.8 | 12 | 70.6 | 6 | 100 | 3 | 100 | 169 | 86.7 |
| Cefepime (FEP) | 120 | 83.9 | 19 | 73.1 | 8 | 47.1 | 6 | 100 | 3 | 100 | 156 | 80 |
| Gentamicin (CN) | 119 | 83.2 | 15 | 57.7 | 17 | 100 | 2 | 33.3 | 0 | 0.0 | 153 | 78.5 |
| Cefotaxime (CTX) | 100 | 69.9 | 25 | 96.2 | 14 | 82.4 | 5 | 83.3 | 1 | 33.3 | 145 | 74.4 |
| Ciprofloxacin (CIP) | 118 | 82.5 | 2 | 7.7 | 17 | 100 | 4 | 66.7 | 3 | 100 | 144 | 73.9 |
| Levofloxacin (LEV) | 73 | 51 | 0 | 0.0 | 8 | 47.1 | 3 | 50 | 1 | 33.3 | 85 | 43.6 |
| Meropenem (MEM) | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Antimicrobial Resistance Patterns * | No. and (%) of Isolates | MAR Index | Resistance Profile | No. and (%) for Each Profile |
|---|---|---|---|---|
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CTX, CIP, LEV | 34 (17.4%) | 0.929 | Extensively drug-resistant | 144 (73.85) |
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, FEP, CTX, CIP, LEV | 6 (3.1%) | 0.857 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP, LEV | 5 (2.6%) | 0.857 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CTX, CIP | 5 (2.6%) | 0.857 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CIP, LEV | 15 (7.7%) | 0.857 | ||
| CEC, FOS, KF, AZM, VA, TE, SXT, FEP, CTX, CIP, LEV | 1 (0.5%) | 0.786 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CTX | 25 (12.8%) | 0.786 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, CN, CTX, CIP, LEV | 5 (2.6%) | 0.786 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP | 14 (7.2%) | 0.786 | ||
| CEC, FOS, KF, NA, VA, TE, SXT, CN, CTX, CIP, LEV | 3 (1.5%) | 0.786 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CIP | 20 (10.3%) | 0.786 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CTX, CIP | 1 (0.5%) | 0.714 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX | 5 (2.6%) | 0.714 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP | 5 (2.6%) | 0.714 | ||
| CEC, FOS, KF, NA, VA, TE, SXT, FEP, CTX, CIP, LEV | 4 (2.1%) | 0.786 | Multidrug-resistant | 51 (26.15%) |
| CEC, FOS, KF, NA, VA, TE, CN, FEP, CTX, CIP, LEV | 1 (0.5%) | 0.786 | ||
| CEC, FOS, KF, NA, AZM, CN, FEP, CTX, CIP, LEV | 10 (5.1%) | 0.714 | ||
| CEC, FOS, KF, NA, AZM, TE, SXT, FEP, CTX, CIP | 5 (2.6%) | 0.714 | ||
| CEC, FOS, KF, NA, AZM, VA, TE, SXT, FEP, CTX | 10 (5.1%) | 0.714 | ||
| CEC, FOS, KF, NA, VA, SXT, CN, CTX, CIP, LEV | 1 (5%) | 0.714 | ||
| CEC, FOS, KF, AZM, VA, TE, SXT, FEP, CIP | 10 (5.1%) | 0.643 | ||
| CEC, FOS, KF, CN, FEP, CTX, CIP | 4 (2.1%) | 0.500 | ||
| CEC, FOS, KF, AZM, VA, CTX | 1 (0.5%) | 0.429 | ||
| CEC, FOS, KF, CN, FEP, CTX | 1 (0.5%) | 0.429 | ||
| CEC, FOS, AZM, VA, CTX | 4 (2.1%) | 0.357 | ||
| Overall Average MAR index = 0.783 | ||||
| Serovars | Number of Isolates | * Antimicrobial Resistance Pattern | Antimicrobial Resistance Classes | MAR Index | Classification | |
|---|---|---|---|---|---|---|
| Type of Resistance | No. and (%) | |||||
| Proteus mirabilis | 31 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.929 | Extensively drug resistant | 103 (78%) |
| 13 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.857 | |||
| 5 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CTX, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.857 | |||
| 3 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.857 | |||
| 3 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.857 | |||
| 16 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 11 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CTX | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides | 0.786 | |||
| 7 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 5 | CEC, FOS, KF, NA, AZM, VA, TE, CN, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 4 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides | 0.714 | |||
| 4 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides | 0.714 | |||
| 1 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CTX, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.714 | |||
| 4 | CEC, FOS, KF, NA, VA, TE, SXT, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.786 | Multidrug-resistant | 29 (22%) | |
| 1 | CEC, FOS, KF, NA, VA, TE, CN, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Glycopeptide, Tetracyclines, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 5 | CEC, FOS, KF, NA, AZM, CN, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Aminoglycosides, Fluoroquinolone | 0.714 | |||
| 5 | CEC, FOS, KF, NA, AZM, TE, SXT, FEP, CTX, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.714 | |||
| 4 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, FEP, CTX | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.714 | |||
| 1 | CEC, FOS, KF, NA, VA, SXT, CN, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Glycopeptide, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.714 | |||
| 6 | CEC, FOS, KF, AZM, VA, TE, SXT, FEP, CIP | Cephalosporin, phosphonic acid, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.643 | |||
| 2 | CEC, FOS, KF, CN, FEP, CTX, CIP | Cephalosporin, phosphonic acid, Aminoglycosides, Fluoroquinolone | 0.500 | |||
| 1 | CEC, FOS, KF, CN, FEP, CTX | Cephalosporin, phosphonic acid, Aminoglycosides | 0.429 | |||
| Sum 132 | Average MAR Index | 0.806 | ||||
| Proteus vulgaris | 3 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.929 | Extensively drug-resistant | 8 (72.7%) |
| 2 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.857 | |||
| 1 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 1 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 1 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides | 0.714 | |||
| 2 | CEC, FOS, KF, NA, AZM, CN, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Aminoglycosides, Fluoroquinolone | 0.714 | Multidrug-resistant | 3 (27.3%) | |
| 1 | CEC, FOS, KF, AZM, VA, TE, SXT, FEP, CIP | Cephalosporin, phosphonic acid, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.643 | |||
| Sum 11 | Average MAR Index | 0.805 | ||||
| Shigella flexneri | 2 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.857 | Extensively drug-resistant | 5 (62.5%) |
| 2 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 1 | CEC, FOS, KF, NA, VA, TE, SXT, CN, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 2 | CEC, FOS, KF, NA, AZM, CN, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Aminoglycosides, Fluoroquinolone | 0.714 | Multidrug-resistant | 3 (37.5%) | |
| 1 | CEC, FOS, KF, CN, FEP, CTX, CIP | Cephalosporin, phosphonic acid, Aminoglycosides, Fluoroquinolone | 0.500 | |||
| Sum 8 | Average MAR Index | 0.750 | ||||
| Shigella sonnei | 1 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | Extensively drug-resistant | 4 (66.7%) |
| 1 | CEC, FOS, KF, NA, VA, TE, SXT, CN, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 2 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 1 | CEC, FOS, KF, NA, AZM, CN, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Aminoglycosides, Fluoroquinolone | 0.714 | Multidrug-resistant | 2 (33.3%) | |
| 1 | CEC, FOS, KF, CN, FEP, CTX, CIP | Cephalosporin, phosphonic acid, Aminoglycosides, Fluoroquinolone | 0.500 | |||
| Sum 6 | Average MAR Index | 0.726 | ||||
| Shigella dysenteriae | 1 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | Extensively drug-resistant | 3 (100%) |
| 1 | CEC, FOS, KF, NA, VA, TE, SXT, CN, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 1 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| Sum 3 | Average MAR Index | 0.786 | ||||
| Citrobacter freundii | 12 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CTX | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides | 0.786 | Extensively drug-resistant | 15 (57.7%) |
| 2 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, CTX, CIP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides, Fluoroquinolone | 0.786 | |||
| 1 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides | 0.714 | |||
| 6 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, FEP, CTX | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides | 0.714 | Multidrug-resistant | 11 (42.3%) | |
| 1 | CEC, FOS, KF, AZM, VA, CTX | Cephalosporin, phosphonic acid, Macrolides, Glycopeptide | 0.429 | |||
| 4 | CEC, FOS, AZM, VA, CTX | Cephalosporin, phosphonic acid, Macrolides, Glycopeptide | 0.357 | |||
| Sum 26 | Average MAR Index | 0.687 | ||||
| Pseudomonas fluorescens | 2 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.857 | Extensively drug-resistant | 5 (83.3%) |
| 2 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, CN, FEP, CTX | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Aminoglycosides | 0.786 | |||
| 1 | CEC, FOS, KF, AZM, VA, TE, SXT, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.786 | |||
| 1 | CEC, FOS, KF, AZM, VA, TE, SXT, FEP, CIP | Cephalosporin, phosphonic acid, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.643 | Multidrug-resistant | 1 (16.7%) | |
| Sum 6 | Average MAR Index | 0.786 | ||||
| Edwardsiella tarda | 1 | CEC, FOS, KF, NA, AZM, VA, TE, SXT, FEP, CTX, CIP, LEV | Cephalosporin, phosphonic acid, Quinolone, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.857 | Extensively drug resistant | 1 (33.3%) |
| 2 | CEC, FOS, KF, AZM, VA, TE, SXT, FEP, CIP | Cephalosporin, phosphonic acid, Macrolides, Glycopeptide, Tetracyclines, Sulfonamides, Fluoroquinolone | 0.643 | Multidrug-resistant | 2 (66.7%) | |
| Sum 3 | Average MAR Index | 0.714 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Saeed, B.A.; Elshebrawy, H.A.; Zakaria, A.I.; Abdelkhalek, A.; Imre, K.; Morar, A.; Herman, V.; Sallam, K.I. Multidrug-Resistant Proteus mirabilis and Other Gram-Negative Species Isolated from Native Egyptian Chicken Carcasses. Trop. Med. Infect. Dis. 2024, 9, 217. https://doi.org/10.3390/tropicalmed9090217
El-Saeed BA, Elshebrawy HA, Zakaria AI, Abdelkhalek A, Imre K, Morar A, Herman V, Sallam KI. Multidrug-Resistant Proteus mirabilis and Other Gram-Negative Species Isolated from Native Egyptian Chicken Carcasses. Tropical Medicine and Infectious Disease. 2024; 9(9):217. https://doi.org/10.3390/tropicalmed9090217
Chicago/Turabian StyleEl-Saeed, Bassant Ashraf, Hend Ali Elshebrawy, Amira Ibrahim Zakaria, Adel Abdelkhalek, Kálmán Imre, Adriana Morar, Viorel Herman, and Khalid Ibrahim Sallam. 2024. "Multidrug-Resistant Proteus mirabilis and Other Gram-Negative Species Isolated from Native Egyptian Chicken Carcasses" Tropical Medicine and Infectious Disease 9, no. 9: 217. https://doi.org/10.3390/tropicalmed9090217
APA StyleEl-Saeed, B. A., Elshebrawy, H. A., Zakaria, A. I., Abdelkhalek, A., Imre, K., Morar, A., Herman, V., & Sallam, K. I. (2024). Multidrug-Resistant Proteus mirabilis and Other Gram-Negative Species Isolated from Native Egyptian Chicken Carcasses. Tropical Medicine and Infectious Disease, 9(9), 217. https://doi.org/10.3390/tropicalmed9090217











