Bovine Leptospirosis in Caatinga Biome, Brazil: New Insights into Diagnosis and Epidemiology
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling
2.3. Biological Sample Collection
2.4. Microscopic Agglutination Test (MAT)
2.5. Microbiological Culture
2.6. Leptospira spp. DNA Detection and Sequencing
2.7. Statistical Analysis
3. Results
3.1. Leptospira spp. Antibody Detection
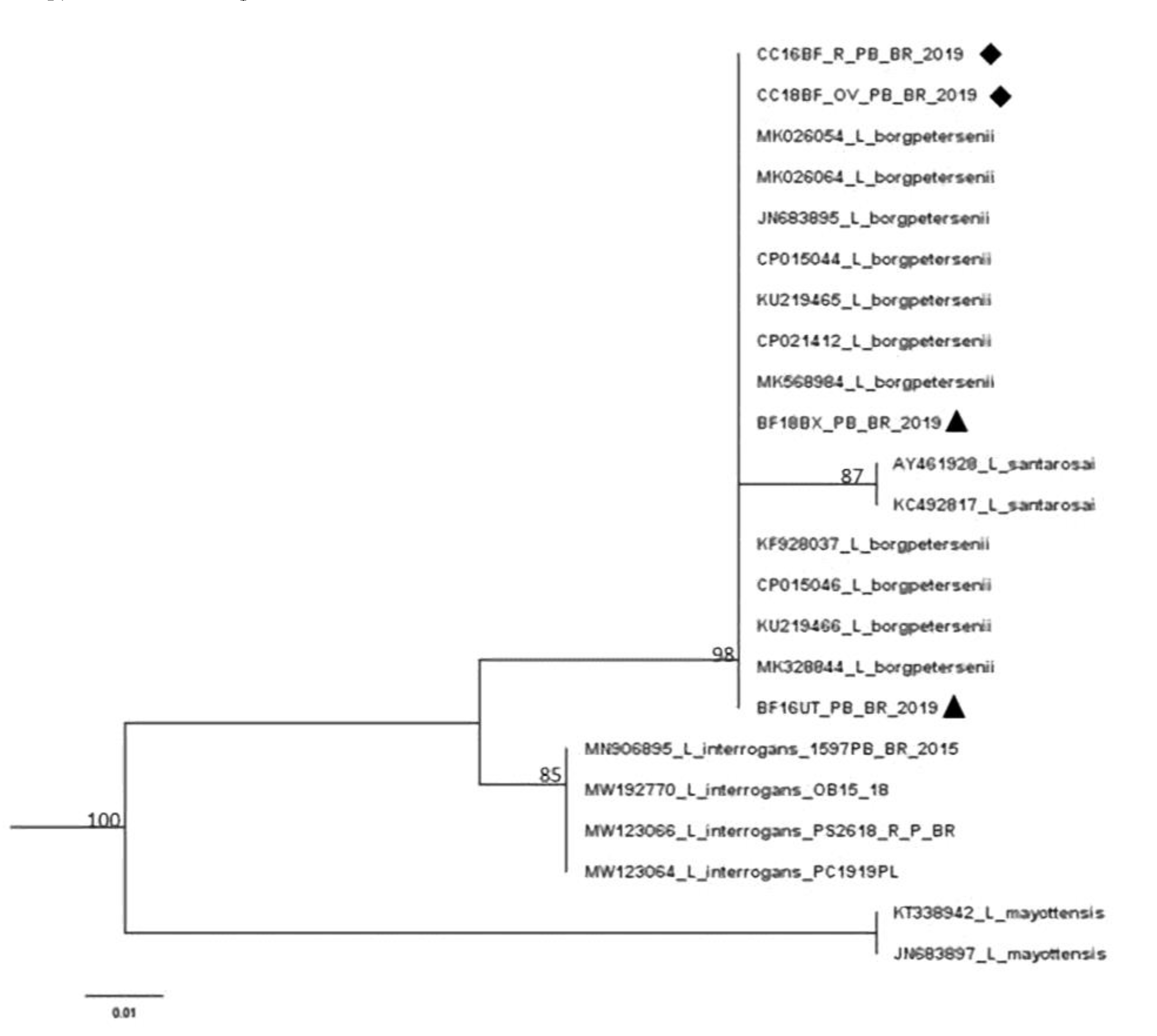
3.2. Leptospira spp. Molecular Results
3.3. Microbiological Culture
3.4. Leptospira spp. DNA Sequencing
3.5. Performance of Diagnostic Tests
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Picardeau, M. Virulence of the zoonotic agent of leptospirosis: Still terra incognita? Nat. Rev. Microbiol. 2017, 15, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global morbidity and mortality of leptospirosis: A systematic review. PLoS Negl. Trop. Dis. 2015, 17, e0003898. [Google Scholar] [CrossRef] [PubMed]
- DE Oliveira, D.; Figueira, C.P.; Zhan, L.; Pertile, A.C.; Pedra, G.G.; Gusmão, I.M.; Wunder, E.A.; Rodrigues, G.; Ramos, E.A.G.; Ko, A.I.; et al. Leptospira in breast tissue and milk of urban Norway rats (Rattus norvegicus). Epidemiol. Infect. 2016, 144, 2420–2429. [Google Scholar] [CrossRef] [PubMed]
- Lilenbaum, W.; Varges, R.; Brandão, F.; Cortez, A.; de Souza, S.; Brandão, P.; Richtzenhain, L.; Vasconcellos, S. Detection of Leptospira spp. in semen and vaginal fluids of goats and sheep by polymerase chain reaction. Theriogenology 2008, 69, 837–842. [Google Scholar] [CrossRef] [PubMed]
- ughini-Gras, L.; Bonfanti, L.; Natale, A.; Comin, A.; Ferronato, A.; LA Greca, E.; Patregnani, T.; Lucchese, L.; Marangon, S. Application of an integrated outbreak management plan for the control of leptospirosis in dairy cattle herds. Epidemiol. Infect. 2014, 142, 1172–1181. [Google Scholar] [CrossRef]
- Loureiro, A.P.; Lilenbaum, W. Genital bovine leptospirosis: A new look for an old disease. Theriogenology 2020, 141, 41–47. [Google Scholar] [CrossRef]
- Pimenta, C.L.R.M.; Castro, V.; Clementino, I.J.; Alves, C.J.; Fernandes, L.G.; Brasil, A.W.L.; Santos, C.S.A.B.; Azevedo, S.S. Bovine leptospirosis in Paraíba State: Prevalence and risk factors associated with the occurrence of positive herds. Pesq. Vet. Bras. 2014, 34, 332–336. [Google Scholar] [CrossRef]
- De Andrade Morais, D.; da Costa, D.F.; Nunes, B.C.; de Sousa Américo Batista Santos, C.; Alves, C.J.; de Azevedo, S.S. Seroepidemiological survey for leptospirosis in equines from semiarid region of Paraíba state, Northeastern Brazil. Semin. Ciências Agrárias 2019, 40, 2079–2086. [Google Scholar] [CrossRef]
- Silva, A.F.; Farias, P.J.A.; Silva, M.L.C.R.; Júnior, J.P.A.; Malossi, C.D.; Ullmann, L.S.; Costa, D.F.; Higino, S.S.S.; Azevedo, S.S.; Alves, C.J. High frequency of genital carriers of Leptospira sp. in sheep slaughtered in the semi-arid region of northeastern Brazil. Trop. Anim. Health Prod. 2019, 51, 43–47. [Google Scholar] [CrossRef]
- Nogueira, D.B.; da Costa, F.T.R.; Bezerra, C.D.S.; Silva, M.L.C.R.; da Costa, D.F.; Viana, M.P.; da Silva, J.D.; Júnior, J.P.A.; Malossi, C.D.; Ullmann, L.S.; et al. Use of serological and molecular techniques for detection of Leptospira sp. carrier sheep under semiarid conditions and the importance of genital transmission route. Acta Trop. 2020, 207, 105497. [Google Scholar] [CrossRef]
- Nogueira, D.B.; da Costa, F.T.R.; Bezerra, C.D.S.; Soares, R.R.; Barnabé, N.N.D.C.; Falcão, B.M.R.; Silva, M.L.C.R.; da Costa, D.F.; Araújo, J.P.; Malossi, C.D.; et al. Leptospira sp. vertical transmission in ewes maintained in semiarid conditions. Anim. Reprod. Sci. 2020, 219, 106530. [Google Scholar] [CrossRef]
- Soares, R.R.; Barnabé, N.N.D.C.; Nogueira, D.B.; da Silva, L.S.C.; Júnior, J.P.A.; Malossi, C.D.; Ullmann, L.S.; da Costa, D.F.; Silva, M.L.C.R.; Higino, S.S.D.S.; et al. Serological, molecular and bacteriological approaches for detecting Leptospira sp. carrier rams maintained in semiarid conditions. Acta Trop. 2021, 213, 105759. [Google Scholar] [CrossRef]
- Fernandes, J.J.; Peixoto, A.D.L.; de Farias, A.S.S.; Pinheiro, T.J.; da Costa, D.F.; Silva, M.L.C.R.; Júnior, J.P.A.; Malossi, C.D.; Ullmann, L.S.; de Azevedo, S.S.; et al. Didelphis albiventris as a carrier of Leptospira sp. in the central nervous tissue in the semiarid region of Northeast, Brazil. Comp. Immunol. Microbiol. Infect. Dis. 2020, 73, 101560. [Google Scholar] [CrossRef]
- Fernandes, J.J.; Pinheiro, T.J.; Costa, D.F.; Júnior, J.P.A.; Malossi, C.D.; Ullmann, L.S.; Silva, M.L.C.R.; Azevedo, S.S.; Alves, C.J.; Higino, S.S.D.S. Leptospira interrogans infection in tegu lizard (Tupinambis merianae), Brazil. Cienc. Rural 2020, 50, e20200424. [Google Scholar] [CrossRef]
- OIE—World Organization for Animal Health. Reference Laboratory Reports Activities; World Organization for Animal Health: Ulster, Northern Ireland, 2014. [Google Scholar]
- Guedes, I.B.; de Souza, G.O.; Castro, J.F.D.P.; Cavalini, M.B.; Filho, A.F.D.S.; Heinemann, M.B. Usefulness of the Ranking Technique in the Microscopic Agglutination Test (MAT) to Predict the Most Likely Infecting Serogroup of Leptospira. Front. Vet. Sci. 2021, 8, 654034. [Google Scholar] [CrossRef]
- Chakraborty, A.; Miyahara, S.; Villanueva, S.Y.A.M.; Saito, M.; Gloriani, N.G.; Yoshida, S.-I. A novel combination of selective agents for isolation of Leptospira species. Microbiol. Immunol. 2011, 55, 494–501. [Google Scholar] [CrossRef]
- Hamond, C.; Martins, G.; Loureiro, A.P.; Pestana, C.; Lawson-Ferreira, R.; Medeiros, M.A.; Lilenbaum, W. Urinary PCR as an increasingly useful tool for an accurate diagnosis of leptospirosis in livestock. Vet. Res. Commun. 2014, 38, 81–85. [Google Scholar] [CrossRef]
- Martins, G.; Lilenbaum, W. Comments of Environmental Conditions for the Maintenance of Leptospira in Tropical Scenarios. Curr. Microbiol. 2015, 71, 624–625. [Google Scholar] [CrossRef]
- Picardeau, M. Diagnosis and epidemiology of leptospirosis. Med. Mal. Infect. 2013, 43, 1–9. [Google Scholar] [CrossRef]
- da Costa, D.F.; da Silva, A.F.; Brasil, A.W.D.L.; Loureiro, A.P.P.; dos Santos, F.A.; de Azevedo, S.S.; Lilenbaum, W.; Alves, C.J. Leptospirosis in native mixed-breed sheep slaughtered in a semiarid region of Brazil. Cienc. Rural 2017, 47, e20160563. [Google Scholar] [CrossRef]
- da Costa, D.F.; Silva, M.L.C.R.; Martins, G.; Dantas, A.F.M.; de Melo, M.A.; de Azevedo, S.S.; Lilenbaum, W.; Alves, C.J. Susceptibility among breeds of sheep experimentally infected with Leptospira interrogans Pomona serogroup. Microb. Pathog. 2018, 122, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Batista, J.S.; Riet-Correa, F.; Teixeira, M.M.G.; Madruga, C.R.; Simões, S.D.V.; Maia, T.F. Trypanosomiasis by Trypanosoma vivax in cattle in the Brazilian semiarid: Description of an outbreak and lesions in the nervous system. Vet. Parasitol. 2007, 143, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Alvares, C.A.; Stape, J.L.; Sentelhas, P.C.; de Moraes Gonçalves, J.L.; Sparovek, G. Köppen’s climate classification map for Brazil. Meteorol. Z. 2014, 22, 711–728. [Google Scholar] [CrossRef] [PubMed]
- INMET—Instituto Nacional de Meteorologia. Estação Meteorológica de Observação de Superfície Automática. Available online: https://www.gov.br/agricultura/pt-br/assuntos/inmet?r=estacoes/estacoesAutomaticas (accessed on 10 May 2021).
- Arango, H.G. Bioestatística Teórica e Computacional, 3rd ed.; Guanabara Koogan: Rio de Janeiro, Brasil, 2009; p. 438. [Google Scholar]
- Pimenta, C.L.R.M.; da Costa, D.F.; Silva, M.L.C.R.; Pereira, H.D.; Araújo Júnior, J.P.; Malossi, C.D.; Ullmann, L.S.; Alves, C.J.; de Azevedo, S.S. Strategies of the control of an outbreak of leptospiral infection in dairy cattle in Northeastern Brazil. Trop. Anim. Health Prod. 2019, 51, 237–241. [Google Scholar] [CrossRef]
- Loureiro, A.P.; Pestana, C.; Medeiros, M.A.; Lilenbaum, W. High frequency of leptospiral vaginal carriers among slaughtered cows. Anim. Reprod. Sci. 2017, 178, 50–54. [Google Scholar] [CrossRef]
- Stoddard, R.A.; Gee, J.E.; Wilkins, P.P.; Mccaustland, K.; Hoffmaster, A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009, 64, 247–255. [Google Scholar] [CrossRef]
- Platt, A.R.; Woodhall, R.W.; George, A.L., Jr. Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. BioTechniques 2007, 43, 58–62. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Ayres, M.; Ayres Junior, M.; Ayres, D.L.; Santos, A.S. Bioestat 5.0 Aplicações Estatísticas nas Áreas das Ciências Biomédicas; ONG Mamiraua: Belém, Brasil, 2007; p. 364. [Google Scholar]
- Mackinnon, A. A spreadsheet for the calculation of comprehensive statistics for the assessment of diagnostic tests and inter-rater agreement. Comput. Biol. Med. 2000, 30, 127–134. [Google Scholar] [CrossRef]
- Pimenta, C.L.R.M.; Nogueira, D.B.; Bezerra, C.D.S.; Morais, D.A.; Silva, M.L.C.R.; Costa, D.F.; Higino, S.S.D.S.; Santos, C.S.A.B.; Alves, C.J.; Azevedo, S.S. High proportion of cattle and sheep seropositive and renal carriers of Leptospira sp. under semiarid conditions. Rev. Bras. Ciênc. Vet. 2020, 27, 22–28. [Google Scholar] [CrossRef]
- Mineiro, A.L.B.B.; Vieira, R.J.; Costa, E.A.; Santos, R.L.; Gonçalves, L.M.F.; Carvalho, S.M.; Bomfim, M.R.Q.; Costa, F.A.L. Serology, polymerase chain reaction and histopathology for leptospirosis in samples collected at slaughter from dairy cows of Parnaiba region, state of Piauí, Brazil. Pesq. Vet. Bras. 2011, 31, 859–866. [Google Scholar] [CrossRef]
- Silva, F.J.; Conceição, W.L.F.; Fagliari, J.J.; Girio, R.J.S.; Dias, R.A.; Borba, M.R.; Mathias, L.A. Prevalence and risk factors of bovine leptospirosis in the State of Maranhão, Brazil. Pesq. Vet. Bras. 2012, 32, 303–312. [Google Scholar] [CrossRef]
- Guedes, I.B.; Araújo, S.A.D.A.; de Souza, G.O.; Silva, S.O.D.S.; Taniwaki, S.A.; Cortez, A.; Brandão, P.E.; Heinemann, M.B. Circulating Leptospira species identified in cattle of the Brazilian Amazon. Acta Trop. 2019, 191, 212–216. [Google Scholar] [CrossRef]
- Guedes, I.B.; de Souza, G.O.; Castro, J.F.D.P.; Filho, A.F.D.S.; Rocha, K.D.S.; Gomes, M.E.T.; de Moraes, C.C.G.; Heinemann, M.B. Development of a pooled antigen for use in the macroscopic slide agglutination test (MSAT) to detect Sejroe serogroup exposure in cattle. J. Microbiol. Methods 2019, 166, 105737. [Google Scholar] [CrossRef]
- Miashiro, A.F.; Vasconcellos, S.A.; de Morais, Z.M.; de Souza, G.O.; Filho, J.M.L.; Figueiredo, A.D.O.; Pellegrin, A.O. Prevalência de leptospirose em rebanhos bovinos no Pantanal de Mato Grosso do Sul. Pesq. Vet. Bras. 2018, 38, 41–47. [Google Scholar] [CrossRef]
- Favero, M.; Pinheiro, S.R.; Vasconcellos, S.A.; Morais, Z.M.; Ferreira, F.; Ferreira Neto, J.S. Bovine leptospirosis—Most frequent serovars in blood collections performed between 1984 to 1997 from herds of 21 brazilian states. Arq. Inst. Biol. 2001, 68, 29–35. [Google Scholar]
- Araújo, V.E.M.; Moreira, E.C.; Naveda, L.A.B.; Silva, J.A.; Contreras, R.L. Frequency of anti-Leptospira interrogans agglutinins in bovine serum samples in Minas Gerais, Brazil, 1980 to 2002. Arq. Bras. Med. Vet. Zootec. 2005, 57, 430–435. [Google Scholar] [CrossRef]
- Pinto, P.S.; Loureiro, A.P.; Penna, B.; Lilenbaum, W. Usage of Leptospira spp. local strains as antigens increases the sensitivity of the serodiagnosis of bovine leptospirosis. Acta Trop. 2015, 149, 163–167. [Google Scholar] [CrossRef]
- Pinna, M.H.; Martins, G.; Loureiro, A.P.; Lilenbaum, W. Detection of bovine carriers of Leptospira by serological, bacteriological, and molecular tools. Trop. Anim. Health Prod. 2018, 50, 883–888. [Google Scholar] [CrossRef]
- Herrmann, G.P.; Rodrigues, R.O.; Machado, G.; Moreira, E.C.; Lage, A.; Leite, R.C. Seroprevalence of leptospirosis in cattle in the southeast and southwest regions of the state of Rio Grande do Sul, Brazil. Ciênc. Anim. Bras. 2012, 13, 131–138. [Google Scholar] [CrossRef]
- Hashimoto, V.Y.; Chideroli, R.T.; Ribeiro, J.; Alfieri, A.A.; Da Costa, G.M.; Pereira, U.D.P.; De Freitas, J.C. Serological and molecular findings in diagnosis of leptospirosis serovar hardjo in a dairy bovine herd. Semin. Cienc. Agrar. 2017, 38, 3155–3164. [Google Scholar] [CrossRef]
- Pinto, P.S.; Pestana, C.; Medeiros, M.A.; Lilenbaum, W. Plurality of Leptospira strains on slaughtered animals suggest a broader concept of adaptability of leptospires to cattle. Acta Trop. 2017, 172, 156–159. [Google Scholar] [CrossRef]
- Ellis, W.A. Animal leptospirosis. Curr. Top. Microbiol. Immunol. 2015, 387, 99–137. [Google Scholar] [CrossRef] [PubMed]
- Soo, Z.M.P.; Khan, N.A.; Siddiqui, R. Leptospirosis: Increasing importance in developing countries. Acta Trop. 2020, 201, 105183. [Google Scholar] [CrossRef]
- Adler, B.; de la Pena Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296. [Google Scholar] [CrossRef]
- Lenharo, D.K.; Santiago, M.E.B.; Lucheis, S.B. Avaliação sorológica para leptospirose em mamíferos silvestres procedentes do parque zoológico municipal de Bauru, SP. Arq. Inst. Biol. 2012, 79, 333–341. [Google Scholar] [CrossRef]
- Hamond, C.; Pestana, C.P.; Medeiros, M.A.; Lilenbaum, W. Genotyping of Leptospira directly in urine samples of cattle demonstrates a diversity of species and strains in Brazil. Epidemiol. Infect. 2016, 144, 72–75. [Google Scholar] [CrossRef]
- Latosinski, G.S.; Fornazari, F.; Babboni, S.D.; Caffaro, K.; Paes, A.C.; Langoni, H. Serological and molecular detection of Leptospira spp. in dogs. Rev. Soc. Bras. Med. Trop. 2018, 51, 364–367. [Google Scholar] [CrossRef]
- Pires, B.C.; Grapiglia, J.B.; Moreira, L.; Jaeger, L.H.; Carvalho-Costa, F.A.; Lilenbaum, W. Occurrence of uterine carriers for Leptospira interrogans on slaughtered cows. Microb. Pathog. 2018, 114, 163–165. [Google Scholar] [CrossRef]
- Zarantonelli, L.; Suanes, A.; Meny, P.; Buroni, F.; Nieves, C.; Salaberry, X.; Briano, C.; Ashfield, N.; Silveira, C.D.S.; Dutra, F.; et al. Isolation of pathogenic Leptospira strains from naturally infected cattle in Uruguay reveals high serovar diversity, and uncovers a relevant risk for human leptospirosis. PLoS Negl. Trop. Dis. 2018, 12, e0006694. [Google Scholar] [CrossRef]
- Merien, F.; Portnoi, D.; Bourhy, P.; Charavay, F.; Berlioz-Arthaud, A.; Baranton, G. A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol. Lett. 2005, 249, 139–147. [Google Scholar] [CrossRef]
- Wynwood, S.J.; Craig, S.B.; Graham, G.C.; Blair, B.R.; Burns, M.A.; Weier, S.L.; Collet, T.A.; McKay, D.B. The emergence of Leptospira borgpetersenii serovar Arborea as the dominant infecting serovar following the summer of natural disasters in Queensland, Australia 2011. Trop. Biomed. 2014, 31, 281–285. [Google Scholar]
- Barragan, V.; Chiriboga, J.; Miller, E.; Olivas, S.; Birdsell, D.; Hepp, C.; Hornstra, H.; Schupp, J.M.; Morales, M.; Gonzalez, M.; et al. High Leptospira Diversity in Animals and Humans Complicates the Search for Common Reservoirs of Human Disease in Rural Ecuador. PLoS Negl. Trop. Dis. 2016, 10, e0004990. [Google Scholar] [CrossRef]
- Grillová, L.; Angermeier, H.; Levy, M.; Giard, M.; Lastère, S.; Picardeau, M. Circulating genotypes of Leptospira in French Polynesia: An 9-year molecular epidemiology surveillance follow-up study. PLoS Negl. Trop. Dis. 2020, 14, e0008662. [Google Scholar] [CrossRef]
- Allan, K.J.; Maze, M.J.; Galloway, R.L.; Rubach, M.P.; Biggs, H.M.; Halliday, J.E.B.; Cleaveland, S.; Saganda, W.; Lwezaula, B.F.; Kazwala, R.R.; et al. Molecular Detection and Typing of Pathogenic Leptospira in Febrile Patients and Phylogenetic Comparison with Leptospira Detected among Animals in Tanzania. Am. J. Trop. Med. Hyg. 2020, 103, 1427–1434. [Google Scholar] [CrossRef]
- Bandara, A.G.N.M.K.; Kalaivarny, G.; Perera, N.; Indrakumar, J. Aseptic meningitis as the initial presentation of Leptospira borgpetersenii serovar Tarassovi: Two case reports and a literature review. BMC Infect. Dis. 2021, 21, 488. [Google Scholar] [CrossRef]
- Grillová, L.; Robinson, M.T.; Chanthongthip, A.; Vincent, A.T.; Nieves, C.; Oppelt, J.; Mariet, J.-F.; Lorioux, C.; Vongsouvath, M.; Mayxay, M.; et al. Genetic diversity of Leptospira isolates in Lao PDR and genome analysis of an outbreak strain. PLoS Negl. Trop. Dis. 2021, 15, e0010076. [Google Scholar] [CrossRef]
- Hernández-Rodríguez, P.; Díaz, C.A.; Dalmau, E.A.; Quintero, G.M. A comparison between polymerase chain reaction (PCR) and traditional techniques for the diagnosis of leptospirosis in bovines. J. Microbiol. Methods 2011, 84, 1–7. [Google Scholar] [CrossRef]
- Koizumi, N.; Micardeau, M. Leptospira spp. Methods and Protocols; Humana: New York, NY, USA, 2020; pp. 277–286. [Google Scholar]
- Ahmed, A.; Engelberts, M.F.M.; Boer, K.R.; Ahmed, N.; Hartskeerl, R.A. Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PLoS ONE 2009, 4, e7093. [Google Scholar] [CrossRef]
| Serogroups | Titers * | Total (%) | |||||
|---|---|---|---|---|---|---|---|
| 50 | 100 | 200 | 400 | 800 | 1600 | ||
| Sejroe | 3 | 5 | 1 | 2 | 2 | 2 | 15 (55.6) |
| Tarassovi | 3 | 1 | 1 | - | - | 1 | 6 (22.2) |
| Australis | 2 | - | - | - | - | - | 2 (7.4) |
| Ballum | 2 | - | - | - | - | - | 2 (7.4) |
| Djasiman | 1 | - | - | - | - | - | 1 (3.7) |
| Hebdomadis | - | - | - | 1 | - | - | 1 (3.7) |
| Total (%) | 11 (40.7) | 6 (22.2) | 2 (7.4) | 3 (11.1) | 2 (7.4) | 3 (11.1) | 27 (100) |
| Sample | Total | PCR | Culture | PCR of Culture | ||
|---|---|---|---|---|---|---|
| 31 */42 (73.8%) | 29 */42 (69%) | 13 */42 (31%) | ||||
| + (%) | Sequencing | + (%) | + (%) | Sequencing | ||
| Urine | 42 | 6 (14.3) a | - | 4 (9.5) a | 1 (2.4) | - |
| Bladder | 42 | 10 (23.8) ab | 1 | 8 (19.1) ab | 1 (2.4) | - |
| Kidney | 42 | 14 (33.3) b | - | 11 (26.2) b | 2 (4.8) | 1 |
| Vaginal fluid | 42 | 10 (23.8) ab | - | 9 (21.4) ab | 1 (2.4) | - |
| Uterus | 42 | 17 (40.5) b | 1 | 13 (31) b | 8 (19.1) | - |
| Uterine tube | 42 | 7 (16.7) a | - | 7 (16.7) ab | 2 (4.8) | - |
| Ovary | 42 | 13 (31) ab | - | 11 (26.2) b | 2 (4.8) | 1 |
| Placenta | 15 | 13 (86.7) c | - | 10 (66.7) c | 2 (13.3) | - |
| Total | 309 | 90 (29.1) | 2 | 73 (23.6) | 19 (6.2) | 2 |
| Biological Material | Microbiological Culture | PCR | ||||
|---|---|---|---|---|---|---|
| 29 */42 (69.05%) | 31 */42 (73.81%) | |||||
| + | - | + | - | SEN | SPE | |
| Urine | 4 | 25 | 6 | 25 | 100 | 94.7 |
| Bladder | 8 | 21 | 10 | 21 | 100 | 94.1 |
| Kidney | 11 | 18 | 14 | 17 | 100 | 90.3 |
| Vaginal fluid | 9 | 20 | 10 | 21 | 100 | 97 |
| Uterus | 13 | 16 | 17 | 14 | 100 | 86.2 |
| Uterine tube | 7 | 22 | 7 | 24 | 100 | 100 |
| Ovary | 11 | 18 | 13 | 18 | 100 | 93.6 |
| Placenta | 10 | 5 | 13 | 2 | 100 | 40 |
| Total | 29 | 13 | 31 | 11 | 100 | 84.6 |
| Biological Material | Antibody Titers (Cut-Off) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 100 | 200 | 400 | |||||||||||||
| 27 */42 (64.3%) | 16 */42 (38.1%) | 10 */42 (23.8%) | 8 */42 (19%) | |||||||||||||
| PCR | MAT | PCR | MAT | PCR | MAT | PCR | MAT | |||||||||
| + | - | SEN | SPE | + | - | SEN | SPE | + | - | SEN | SPE | + | - | SEN | SPE | |
| Urine | 4 | 23 | 66.7 | 36.1 | 3 | 13 | 50 | 63.9 | 2 | 8 | 33.3 | 77.8 | 2 | 6 | 33.3 | 83.3 |
| Bladder | 7 | 20 | 70 | 37.5 | 5 | 11 | 50 | 65.6 | 4 | 6 | 40 | 81.3 | 3 | 5 | 30 | 84.4 |
| Kidney | 9 | 18 | 64.3 | 35.7 | 6 | 10 | 42.9 | 64.3 | 5 | 5 | 35.7 | 82.1 | 4 | 4 | 28.6 | 85.7 |
| Vaginal fluid | 6 | 21 | 60 | 34.4 | 5 | 11 | 50 | 65.6 | 3 | 7 | 30 | 78.1 | 3 | 5 | 30 | 84.4 |
| Uterus | 8 | 19 | 47.1 | 24 | 6 | 10 | 35.3 | 60 | 4 | 6 | 23.5 | 76 | 4 | 4 | 23.5 | 84 |
| Uterine tube | 4 | 23 | 57.1 | 34.3 | 1 | 15 | 14.3 | 57.1 | 1 | 9 | 14.3 | 74.3 | 1 | 7 | 14.3 | 80 |
| Ovary | 8 | 19 | 61.5 | 34.5 | 3 | 13 | 23.1 | 55.2 | 3 | 7 | 23.1 | 75.9 | 3 | 5 | 23.1 | 82.8 |
| Placenta | 7 | 2 | 53.9 | 0 | 5 | 1 | 38.5 | 50 | 3 | 1 | 23.1 | 50 | 3 | 1 | 23.1 | 50 |
| Biological Material | Antibody Titers (Cut-Off) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 100 | 200 | 400 | |||||||||||||
| 27 */42 (64.3%) | 16 */42 (38.1%) | 10 */42 (23.8%) | 8 */42 (19%) | |||||||||||||
| MC | MAT | MC | MAT | MC | MAT | MC | MAT | |||||||||
| + | - | SEN | SPE | + | - | SEN | SPE | + | - | SEN | SPE | + | - | SEN | SPE | |
| Urine | 4 | 23 | 100 | 39.5 | 3 | 13 | 75 | 65.8 | 2 | 8 | 50 | 79 | 2 | 6 | 50 | 84.2 |
| Bladder | 6 | 21 | 75 | 38.2 | 4 | 12 | 50 | 64.7 | 3 | 7 | 37.5 | 79.4 | 3 | 5 | 37.5 | 85.3 |
| Kidney | 8 | 19 | 72 | 38.7 | 5 | 11 | 45.5 | 64.5 | 5 | 5 | 45.5 | 83.9 | 4 | 4 | 36.4 | 87.1 |
| Vaginal fluid | 6 | 21 | 66.7 | 36.4 | 5 | 11 | 55.6 | 66.7 | 3 | 7 | 33.3 | 78.8 | 3 | 5 | 33.3 | 84.9 |
| Uterus | 7 | 20 | 53.9 | 31 | 5 | 11 | 38.5 | 62.1 | 3 | 7 | 23.1 | 75.9 | 3 | 5 | 23.1 | 82.8 |
| Uterine tube | 4 | 23 | 57.1 | 34.3 | 1 | 15 | 14.3 | 57.1 | 1 | 9 | 14.3 | 74.3 | 1 | 7 | 14.3 | 80 |
| Ovary | 7 | 20 | 63.6 | 35.5 | 3 | 13 | 27.3 | 58.1 | 3 | 7 | 27.3 | 77.4 | 3 | 5 | 27.3 | 83.9 |
| Placenta | 6 | 3 | 60 | 40 | 4 | 2 | 40 | 60 | 2 | 2 | 20 | 60 | 2 | 2 | 20 | 60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barnabé, N.N.d.C.; Soares, R.R.; Barros, D.K.S.; Nogueira, D.B.; Costa, F.T.R.d.; Araújo Júnior, J.P.; Malossi, C.D.; Ullmann, L.S.; Costa, D.F.d.; Silva, M.L.C.R.; et al. Bovine Leptospirosis in Caatinga Biome, Brazil: New Insights into Diagnosis and Epidemiology. Trop. Med. Infect. Dis. 2023, 8, 177. https://doi.org/10.3390/tropicalmed8030177
Barnabé NNdC, Soares RR, Barros DKS, Nogueira DB, Costa FTRd, Araújo Júnior JP, Malossi CD, Ullmann LS, Costa DFd, Silva MLCR, et al. Bovine Leptospirosis in Caatinga Biome, Brazil: New Insights into Diagnosis and Epidemiology. Tropical Medicine and Infectious Disease. 2023; 8(3):177. https://doi.org/10.3390/tropicalmed8030177
Chicago/Turabian StyleBarnabé, Nathanael Natércio da Costa, Rafael Rodrigues Soares, Deivyson Kelvis Silva Barros, Denise Batista Nogueira, Flávia Teresa Ribeiro da Costa, João Pessoa Araújo Júnior, Camila Dantas Malossi, Leila Sabrina Ullmann, Diego Figueiredo da Costa, Maria Luana Cristiny Rodrigues Silva, and et al. 2023. "Bovine Leptospirosis in Caatinga Biome, Brazil: New Insights into Diagnosis and Epidemiology" Tropical Medicine and Infectious Disease 8, no. 3: 177. https://doi.org/10.3390/tropicalmed8030177
APA StyleBarnabé, N. N. d. C., Soares, R. R., Barros, D. K. S., Nogueira, D. B., Costa, F. T. R. d., Araújo Júnior, J. P., Malossi, C. D., Ullmann, L. S., Costa, D. F. d., Silva, M. L. C. R., Higino, S. S. d. S., Santos, C. d. S. A. B., Azevedo, S. S. d., & Alves, C. J. (2023). Bovine Leptospirosis in Caatinga Biome, Brazil: New Insights into Diagnosis and Epidemiology. Tropical Medicine and Infectious Disease, 8(3), 177. https://doi.org/10.3390/tropicalmed8030177









