Biomphalaria pfeifferi (Gastropoda: Planorbidae) in Lake Malawi and Upper Shire River, Mangochi District, Malawi: Distribution, Genetic Diversity and Pre-Patent Schistosome Infections
Abstract
1. Introduction
2. Materials and Methods
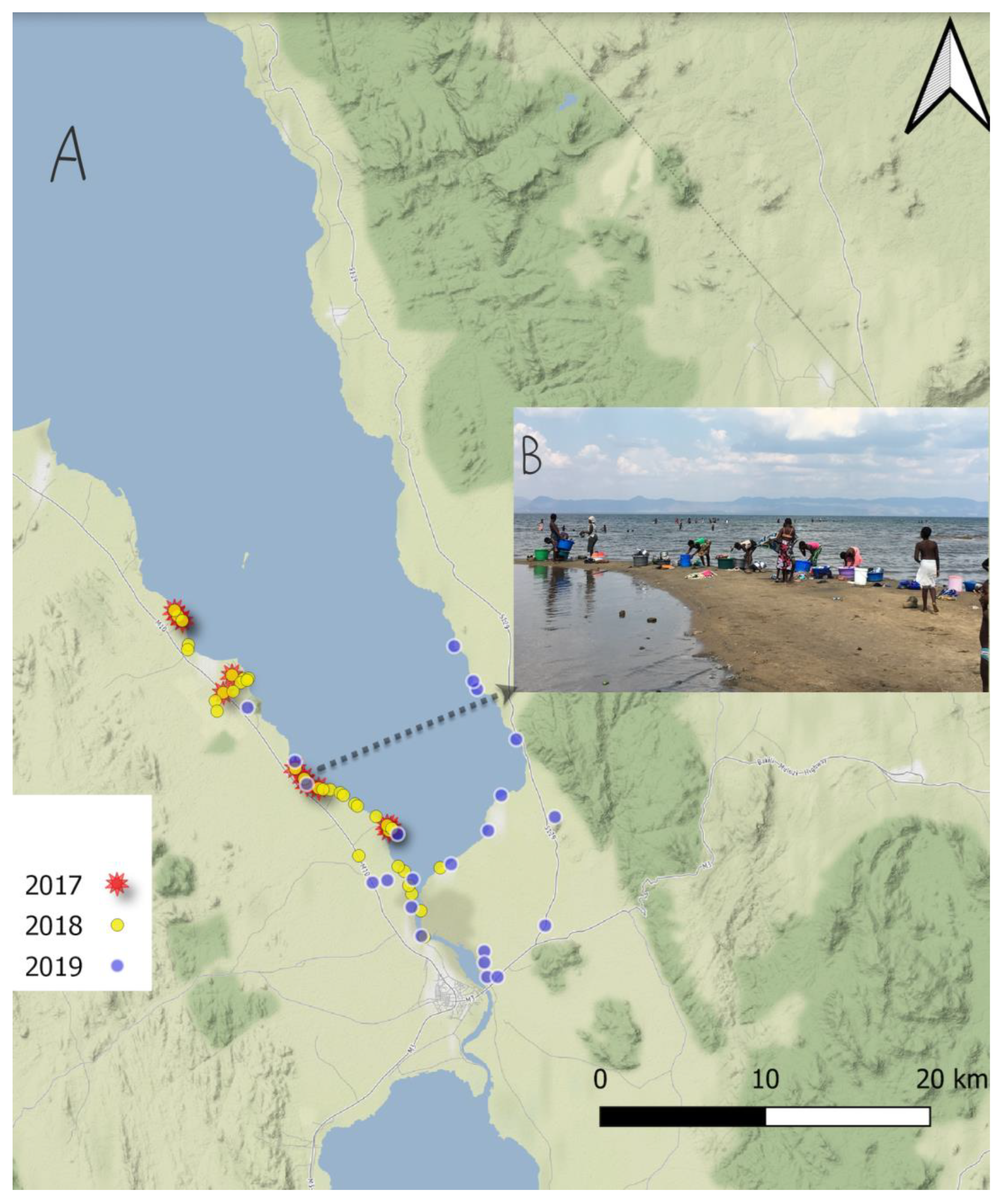
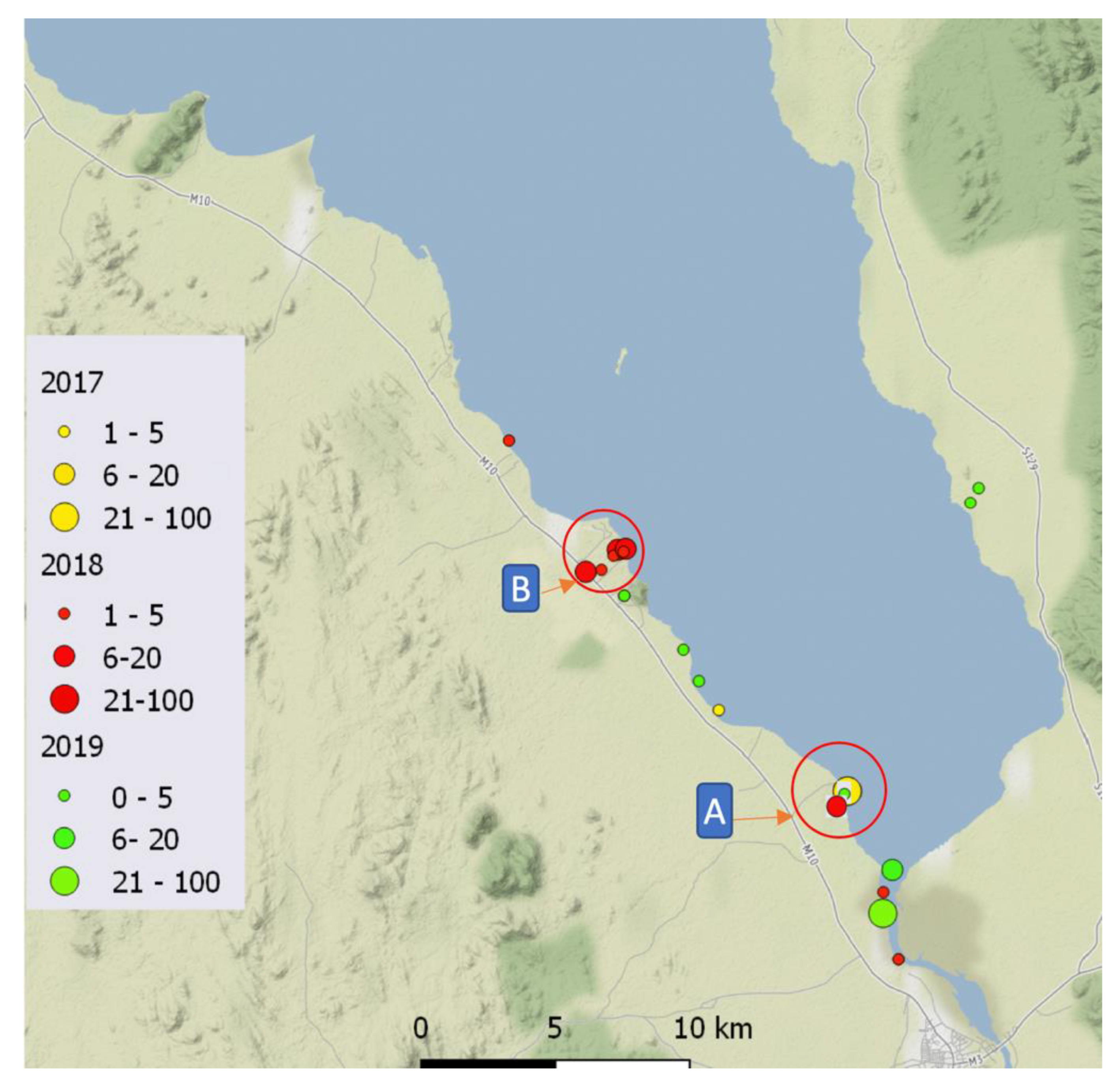
2.1. Malacological Surveillance
2.2. Morphological Analyses, Sample Preparation and DNA Extraction
2.3. PCR and DNA Sequencing Analysis
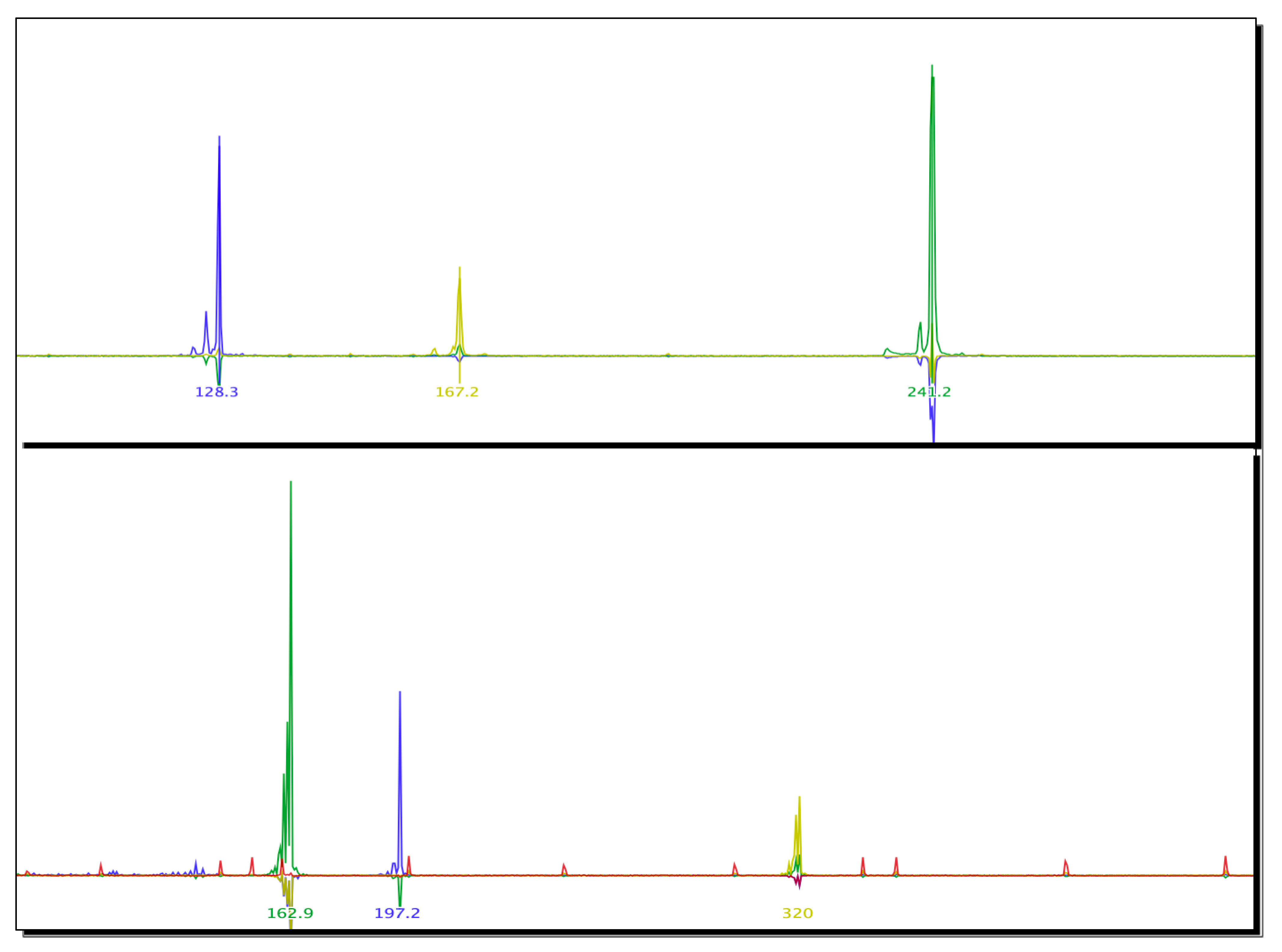
2.4. Microsatellite Markers Used to Characterize Biomphalaria spp. Specimens
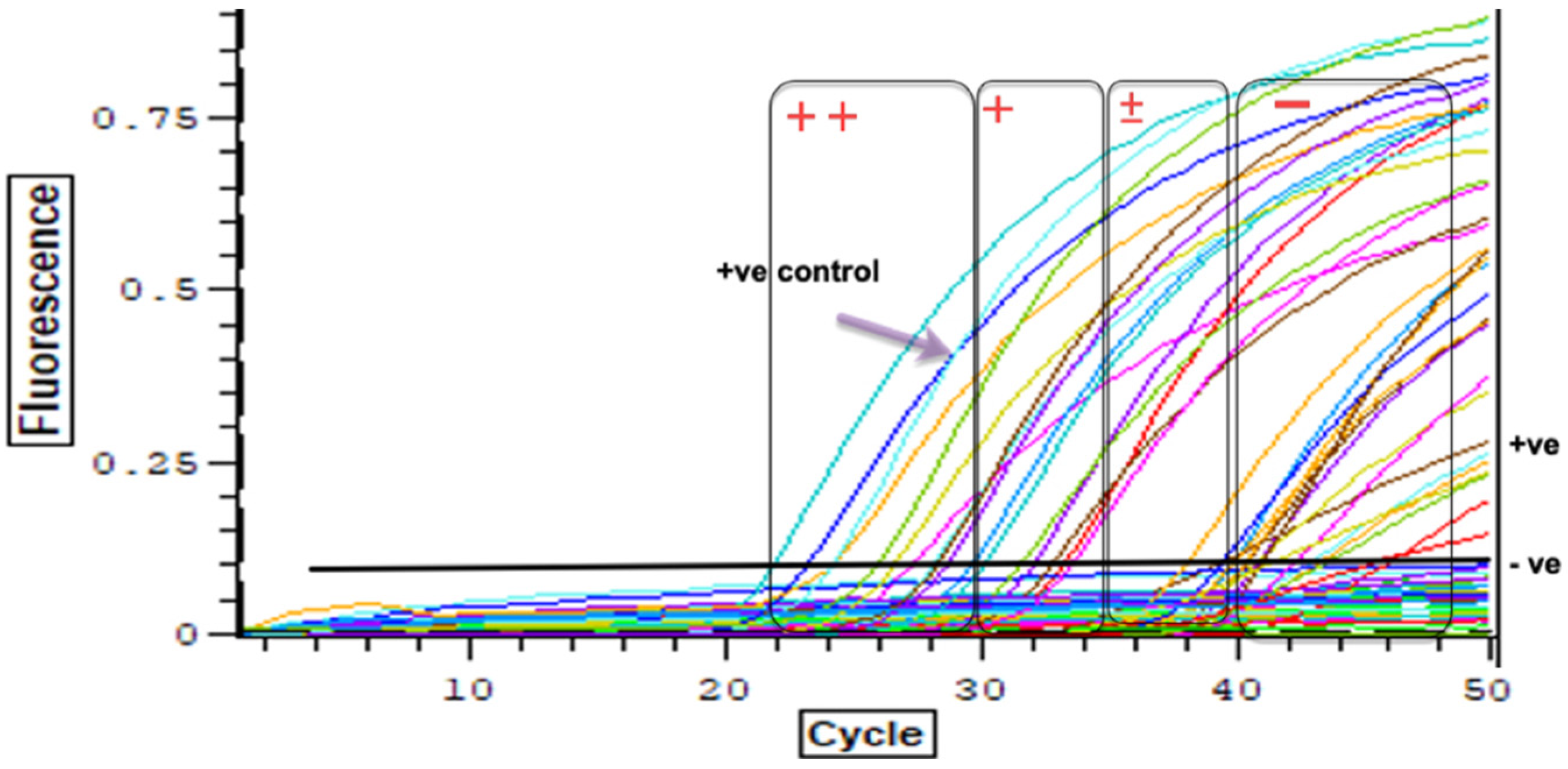
2.5. Real-Time PCR to Detect Pre-Patent Schistosoma spp. Infections within Snails
2.6. Data Handling and Statistical Analysis
3. Results
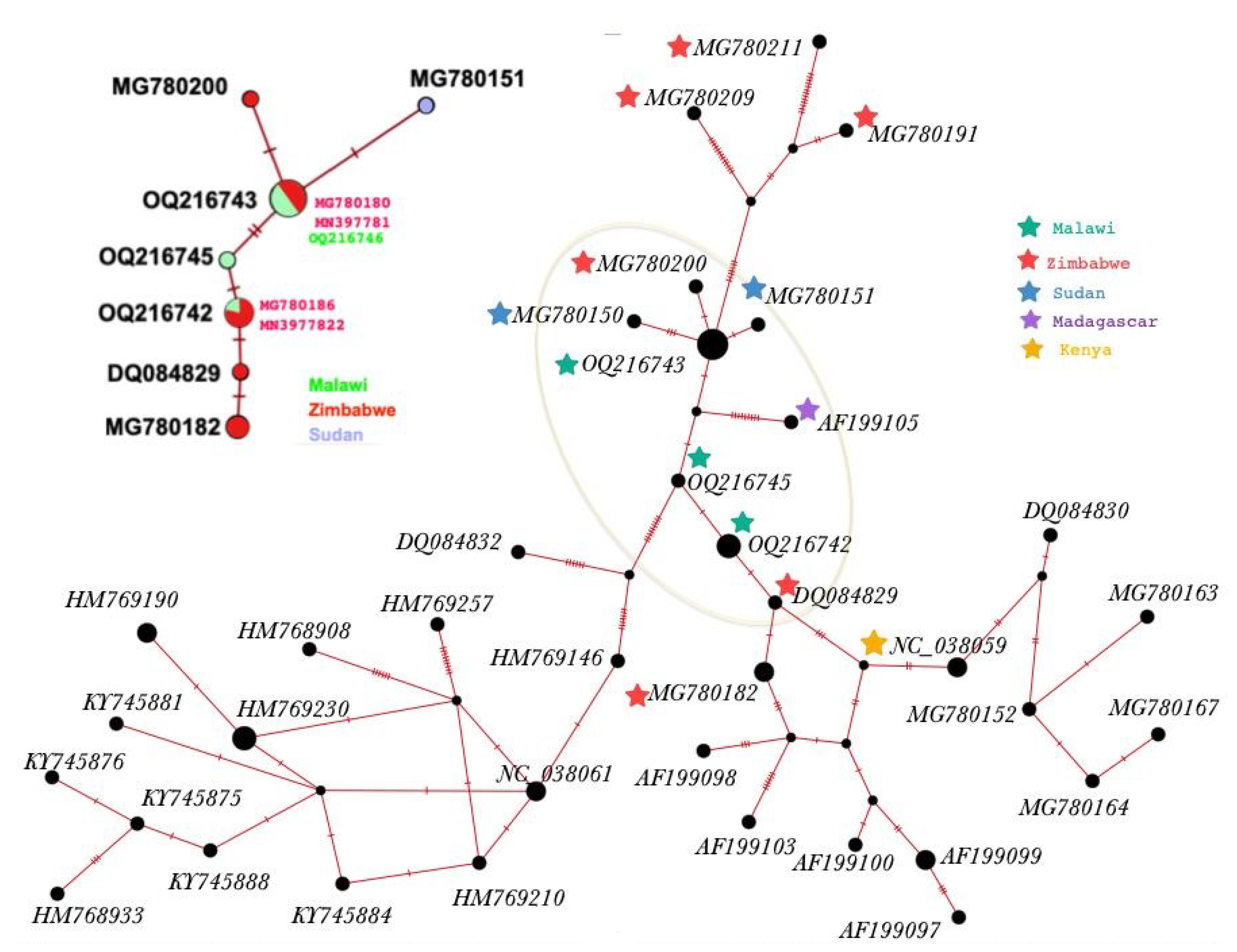
3.1. Molecular Characterisation
3.2. Microsatellite Data Analysis
3.3. Detection of Schistosoma spp. DNA within Collected Bi. pfeifferi Specimens
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Brown, D.S. Freshwater Snails of Africa and Their Medical Importance; CRC Press: London, UK, 1994. [Google Scholar]
- Stensgaard, A.-S.; Utzinger, J.; Vounatsou, P.; Hürlimann, E.; Schur, N.; Saarnak, C.F.; Simoonga, C.; Mubita, P.; Kabatereine, N.B.; Tchuenté, L.-A.T. Large-scale determinants of intestinal schistosomiasis and intermediate host snail distribution across Africa: Does climate matter? Acta Trop. 2013, 128, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Madsen, H.; Stauffer, J.R. Schistosomiasis Control Under Changing Ecological Settings in Lake Malawi. EcoHealth 2022, 19, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Cetron, M.S.; Chitsulo, L.; Sullivan, J.J.; Pilcher, J.; Wilson, M.; Noh, J.; Tsang, V.C.; Hightower, A.W.; Addiss, D.G. Schistosomiasis in lake Malawi. Lancet 1996, 348, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Chidammodzi, C.; Muhandiki, V. Development of indicators for assessment of Lake Malawi Basin in an Integrated Lake Basin Management (ILBM) framework. Int. J. Commons 2015, 9, 209–236. [Google Scholar] [CrossRef]
- Stauffer, J.R.; Arnegard, M.E.; Cetron, M.; Sullivan, J.J.; Chitsulo, L.A.; Turner, G.F.; Chiotha, S.; McKaye, K. Controlling vectors and hosts of parasitic diseases using fishes. BioScience 1997, 47, 41–49. [Google Scholar] [CrossRef]
- Bootsma, H.A.; Jorgensen, S.E. Lake Malawi/Nyasa: Experience and Lessons Learned Brief. 2013. Available online: https://www.semanticscholar.org/paper/Lake-Malawi%2FNyasa%3A-Experience-and-lessons-learned-Bootsma-J%C3%B8rgensen/4eeb195d7d53a1fe2f8afc8ee77b310503d79804 (accessed on 12 December 2022).
- Alharbi, M.H.; Iravoga, C.; Kayuni, S.A.; Cunningham, L.; LaCourse, E.J.; Makaula, P.; Stothard, J.R. First Molecular Identification of Bulinus africanus in Lake Malawi Implicated in Transmitting Schistosoma Parasites. Trop. Med. Infect. Dis. 2022, 7, 195. [Google Scholar] [CrossRef]
- Webster, B.L.; Alharbi, M.; Kayuni, S.; Makaula, P.; Halstead, F.; Christiansen, R.; Juziwelo, L.; Stanton, M.; LaCourse, J.; Rollinson, D.; et al. Schistosome Interactions within the Schistosoma Haematobium Group, Malawi; National Center for Infectious Diseases: Atlanta, GA, USA, 2019. [Google Scholar]
- Alharbi, M.H.; Condemine, C.; Christiansen, R.; LaCourse, E.J.; Makaula, P.; Stanton, M.C.; Juziwelo, L.; Kayuni, S.; Stothard, J.R. Biomphalaria pfeifferi snails and intestinal schistosomiasis, Lake Malawi, Africa, 2017–2018. Emerg. Infect. Dis. 2019, 25, 613. [Google Scholar] [CrossRef]
- Kayuni, S.A.; O’Ferrall, A.M.; Baxter, H.; Hesketh, J.; Mainga, B.; Lally, D.; Al-Harbi, M.H.; LaCourse, E.J.; Juziwelo, L.; Musaya, J. An outbreak of intestinal schistosomiasis, alongside increasing urogenital schistosomiasis prevalence, in primary school children on the shoreline of Lake Malawi, Mangochi District, Malawi. Infect. Dis. Poverty 2020, 9, 121. [Google Scholar] [CrossRef]
- Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Charbonnel, N.; Angers, B.; Razatavonjizay, R.; Bremond, P.; Jarne, P. Microsatellite variation in the freshwater snail Biomphalaria pfeifferi. Mol. Ecol. 2000, 9, 1006–1007. [Google Scholar] [CrossRef]
- Nguema, R.M.; Langand, J.; Galinier, R.; Idris, M.A.; Shaban, M.A.; Al Yafae, S.; Moné, H.; Mouahid, G. Genetic diversity, fixation and differentiation of the freshwater snail Biomphalaria pfeifferi (Gastropoda, Planorbidae) in arid lands. Genetica 2013, 141, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.S.; Lockyer, A.E.; Rollinson, D.; Piertney, S.B.; Noble, L.R. Isolation and characterization of microsatellite loci in the freshwater gastropod, Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Mol. Ecol. 1999, 8, 2149–2151. [Google Scholar] [CrossRef]
- Habib, M.R.; Lv, S.; Rollinson, D.; Zhou, X.-N. Invasion and Dispersal of Biomphalaria Species: Increased Vigilance Needed to Prevent the Introduction and Spread of Schistosomiasis. Front. Med. 2021, 8, 614797. [Google Scholar] [CrossRef] [PubMed]
- Angers, B.; Charbonnel, N.; Galtier, N.; Jarne, P. The influence of demography, population structure and selection on molecular diversity in the selfing freshwater snail Biomphalaria pfeifferi. Genet. Res. 2003, 81, 193–204. [Google Scholar] [CrossRef]
- Webster, J.; Davies, C.; Hoffman, J.; Ndamba, J.; Noble, L.; Woolhouse, M. Population genetics of the schistosome intermediate host Biomphalaria pfeifferi in the Zimbabwean highveld: Implications for co-evolutionary theory. Ann. Trop. Med. Parasitol. 2001, 95, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Sandland, G.J.; Wethington, A.R.; Foster, A.V.; Minchella, D.J. Effects of host outcrossing on the interaction between an aquatic snail and its locally adapted parasite. Parasitol. Res. 2009, 105, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Sandland, G.J.; Foster, A.V.; Zavodna, M.; Minchella, D.J. Interplay between host genetic variation and parasite transmission in the Biomphalaria glabrata–Schistosoma mansoni system. Parasitol. Res. 2007, 101, 1083–1089. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, B.G. Schistosoma: Biology, Pathology and Control; CRC Press: London, UK, 2017. [Google Scholar]
- Pennance, T.; Archer, J.; Lugli, E.B.; Rostron, P.; Llanwarne, F.; Ali, S.M.; Amour, A.K.; Suleiman, K.R.; Li, S.; Rollinson, D. Development of a molecular snail xenomonitoring assay to detect Schistosoma haematobium and Schistosoma bovis infections in their Bulinus snail hosts. Molecules 2020, 25, 4011. [Google Scholar] [CrossRef]
- Kamel, B.; Laidemitt, M.R.; Lu, L.; Babbitt, C.; Weinbaum, O.L.; Mkoji, G.M.; Loker, E.S. Detecting and identifying Schistosoma infections in snails and aquatic habitats: A systematic review. PLoS Negl. Trop. Dis. 2021, 15, e0009175. [Google Scholar] [CrossRef]
- Rollinson, D.; Knopp, S.; Levitz, S.; Stothard, J.R.; Tchuenté, L.-A.T.; Garba, A.; Mohammed, K.A.; Schur, N.; Person, B.; Colley, D.G. Time to set the agenda for schistosomiasis elimination. Acta Trop. 2013, 128, 423–440. [Google Scholar] [CrossRef]
- Born-Torrijos, A.; Poulin, R.; Raga, J.A.; Holzer, A.S. Estimating trematode prevalence in snail hosts using a single-step duplex PCR: How badly does cercarial shedding underestimate infection rates? Parasites Vectors 2014, 7, 243. [Google Scholar] [CrossRef] [PubMed]
| Locus | Primer Sequence (5′-3′) | Source |
|---|---|---|
| Bpf8 | F:GGTTCCCATAGCATACAGTGC R:GGCTTACAAAGAAACAGGCATAC | [13] |
| U-7 | F: TCATGACATCTAATGGGAGAAG R:GGGCTCAGAGAAATGAATGG | Unpublished data |
| rg6 | F: GATTTTGTCTCACGGAAACG R:GCGTGCTTATGTAGCAAAGG | [14] |
| rg9 | F:GGAGCTGTCGTAATTATAGTTCA R:TTGAGGTGTCATGGTTCTAGG | [14] |
| Bgµ16 | F:CTGTTATTCATTATTTCATAGAGC R: GGGGATCTAACACATCAG | [15] |
| µBg1 | F:TTAATTCTACTGGACTCACATGG R:CTGCCAATGTTTACATGCTG | [15] |
| Haplotype & Acc.No | 169 | 187 | 250 | 537 | 569 |
|---|---|---|---|---|---|
| NC_038059 | T | G | A | G | G |
| H1 (OQ216742) | T | G | A | G | G |
| H2 (OQ216743) | C | A | G | G | G |
| H3 (OQ216744) | C | A | G | T | G |
| H4 (OQ216745) | T | A | A | G | G |
| H5 (OQ216746) | C | A | G | G | T |
| N | Marker | Expected Amplicon Size Range (bp) | Size of Amplicon | Allele Frequency |
|---|---|---|---|---|
| 1 | Bgμl6 | 113–135 | 128 bp | 1.00 |
| 2 | Bpf8 | 193–225 | 167 bp | 1.00 |
| 3 | rg6 | 233–244 | 241 bp | 1.00 |
| 4 | μBG1 | 165–167 | 162 bp | 1.00 |
| 5 | U7 | 222–246 | 197 bp | 1.00 |
| 6 | RG9 | 313–321 | 320 bp | 1.00 |
| Location | Type | Year | Latitude | Longitude | Number of Snails Examined | Infected | Haplotype and Acc.No |
|---|---|---|---|---|---|---|---|
| 1 | Lake | 2017 | −14.36919 | 35.17629 | 2 | 0 | H1 OQ216742 |
| 2 | Lake | −14.39363 | 35.22104 | 17 | 2 | H1 OQ216742 H2 OQ216743 H3 OQ216744 H4 OQ216745 H5 OQ216746 | |
| 3 | Lake | 2018 | −14.27752 | 35.10419 | 1 | 0 | H1 OQ216742 |
| 4 | Stream | −14.32120 | 35.13072 | 5 | 4 | ||
| 5 | Lake | −14.39363 | 35.22104 | 22 | 5 | ||
| 6 | Lake | −14.44975 | 35.24018 | 3 | 3 | ||
| 7 | Lake | −14.42708 | 35.23349 | 1 | 0 | ||
| 8 | Stream | −14.31371 | 35.14174 | 4 | 4 | ||
| 9 | Stream | −14.31568 | 35.14030 | 2 | 2 | ||
| 10 | Stream | −14.32033 | 35.13613 | 2 | 1 | ||
| 11 | Stream | −14.31338 | 35.14449 | 1 | 0 | ||
| 12 | Lake | −14.31437 | 35.14376 | 2 | 0 | ||
| 13 | Lake | 2019 | −14.44930 | 35.23871 | 1 | 0 | |
| 14 | River | −14.34131 | 35.28897 | 2 | 1 | ||
| 15 | River | −14.41937 | 35.23394 | 2 | 1 | ||
| 16 | River | −14.32895 | 35.14481 | 1 | 1 | ||
| 17 | Lake | −14.39545 | 35.22569 | 3 | 0 | ||
| 18 | Lake | −14.29649 | 35.25654 | 2 | 0 | ||
| 19 | Lake | −14.31494 | 35.26697 | 2 | 0 | ||
| 20 | Lake | −14.36913 | 35.17623 | 1 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alharbi, M.H.; Condemine, C.; Hesketh, J.; Kayuni, S.A.; Arme, T.M.; Archer, J.; Jones, S.; LaCourse, E.J.; Makaula, P.; Musaya, J.; et al. Biomphalaria pfeifferi (Gastropoda: Planorbidae) in Lake Malawi and Upper Shire River, Mangochi District, Malawi: Distribution, Genetic Diversity and Pre-Patent Schistosome Infections. Trop. Med. Infect. Dis. 2023, 8, 126. https://doi.org/10.3390/tropicalmed8020126
Alharbi MH, Condemine C, Hesketh J, Kayuni SA, Arme TM, Archer J, Jones S, LaCourse EJ, Makaula P, Musaya J, et al. Biomphalaria pfeifferi (Gastropoda: Planorbidae) in Lake Malawi and Upper Shire River, Mangochi District, Malawi: Distribution, Genetic Diversity and Pre-Patent Schistosome Infections. Tropical Medicine and Infectious Disease. 2023; 8(2):126. https://doi.org/10.3390/tropicalmed8020126
Chicago/Turabian StyleAlharbi, Mohammad H., Charlotte Condemine, Josie Hesketh, Sekeleghe A. Kayuni, Thomas M. Arme, John Archer, Sam Jones, E. James LaCourse, Peter Makaula, Janelisa Musaya, and et al. 2023. "Biomphalaria pfeifferi (Gastropoda: Planorbidae) in Lake Malawi and Upper Shire River, Mangochi District, Malawi: Distribution, Genetic Diversity and Pre-Patent Schistosome Infections" Tropical Medicine and Infectious Disease 8, no. 2: 126. https://doi.org/10.3390/tropicalmed8020126
APA StyleAlharbi, M. H., Condemine, C., Hesketh, J., Kayuni, S. A., Arme, T. M., Archer, J., Jones, S., LaCourse, E. J., Makaula, P., Musaya, J., & Stothard, J. R. (2023). Biomphalaria pfeifferi (Gastropoda: Planorbidae) in Lake Malawi and Upper Shire River, Mangochi District, Malawi: Distribution, Genetic Diversity and Pre-Patent Schistosome Infections. Tropical Medicine and Infectious Disease, 8(2), 126. https://doi.org/10.3390/tropicalmed8020126






