Polymorphism of Drug Resistance Genes dhfr and dhps in Plasmodium falciparum Isolates among Chinese Migrant Workers Who Returned from Ghana in 2013
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Considerations
2.2. Sample Collection and DNA Extraction
2.3. PCR Amplification and Sequencing
2.4. Chronological Analysis of SP Resistance
3. Results
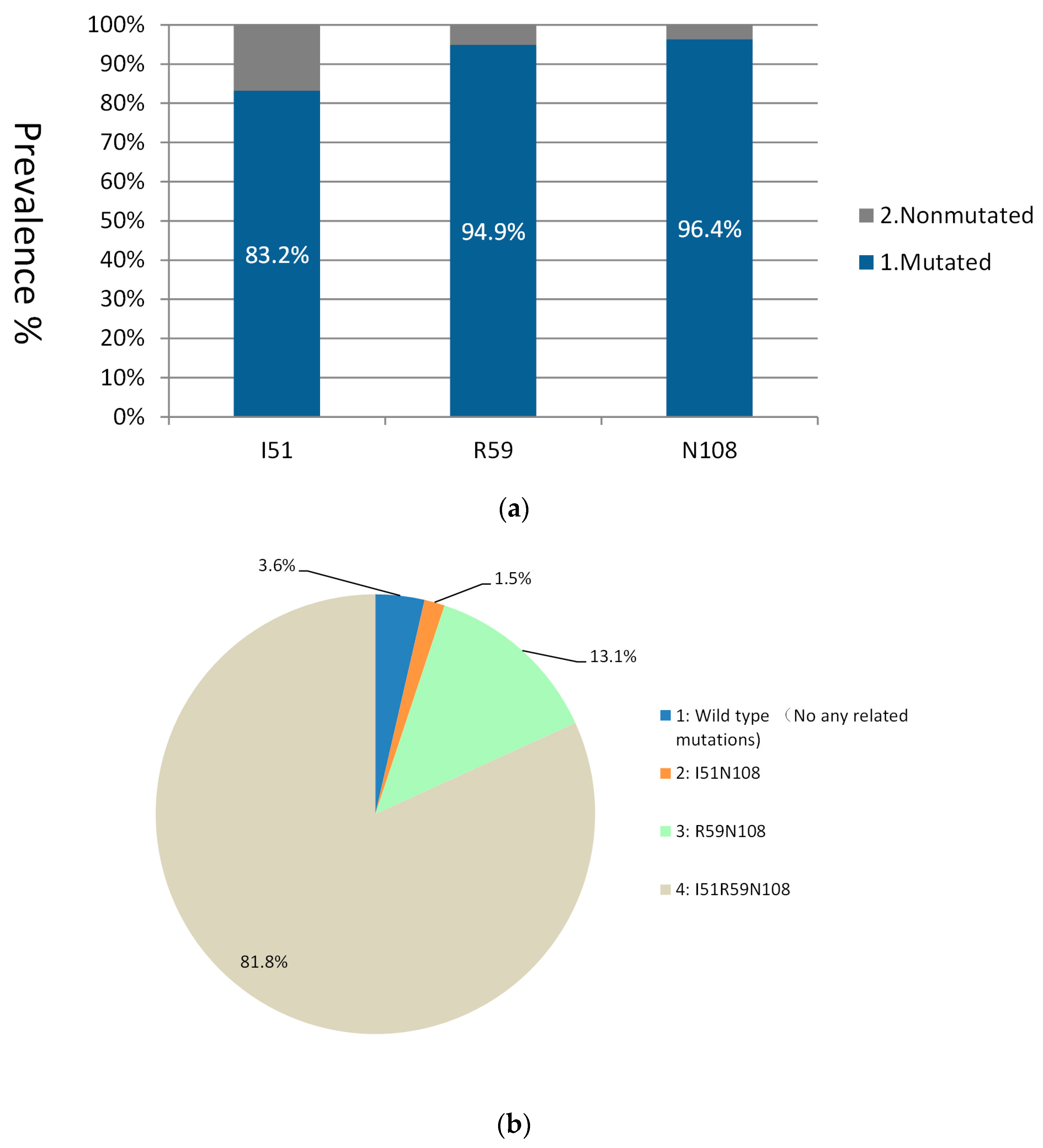
3.1. Mutations in the dhfr Gene
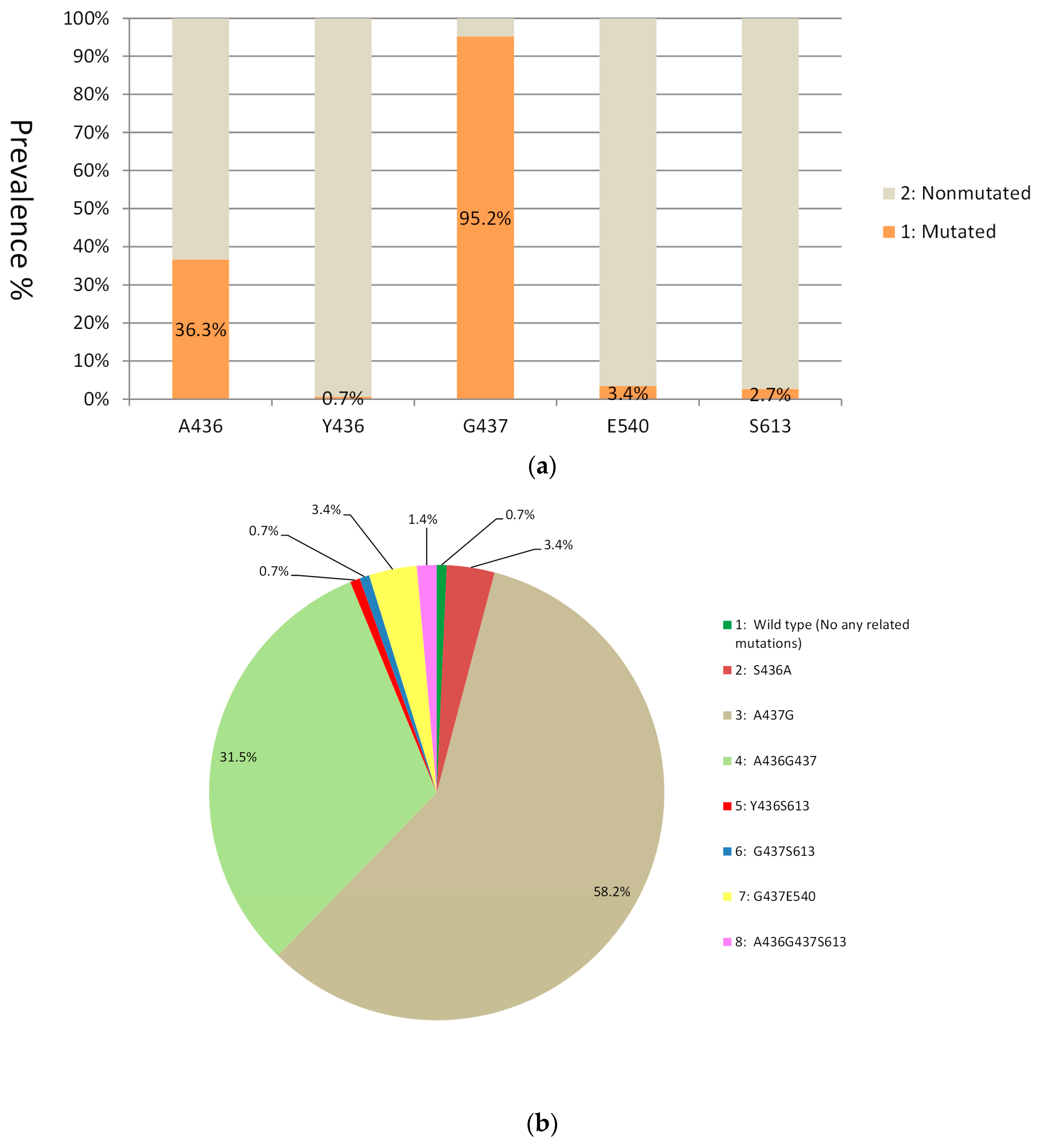
3.2. Mutations in the dhps Gene
3.3. dhfr/dhps Haplotype Combination
3.4. Chronological Analysis of SP Resistance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. World Malaria Report 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Hu, T.; Liu, Y.-B.; Zhang, S.-S.; Xia, Z.-G.; Zhou, S.-S.; Yan, J.; Cao, J.; Feng, Z.-C. Shrinking the malaria map in China: Measuring the progress of the National Malaria Elimination Programme. Infect. Dis. Poverty 2016, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, L.; Zhang, S.S.; Xia, Z.G.; Zhou, S.S. Malaria epidemiological characteristics in China, 2005–2015. CTM 2017, 17, 325–335. [Google Scholar]
- Zhou, H.-Y.; Wang, W.-M.; Zhu, G.-D.; Cao, Y.-Y.; Lu, F.; Gu, Y.-P.; Zhang, C.; Xu, S.; Cao, J. Epidemiological analysis of malaria prevalence in Jiangsu Province in 2016. Chin. J. Schistosomiasis Control 2018, 30, 32–36. [Google Scholar]
- Liu, Y.; Sturrock, H.J.W.; Yang, H.; Gosling, R.D.; Cao, J. The challenge of imported malaria to eliminating countries. Lancet Infect. Dis. 2017, 17, 141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Feng, J.; Zhang, S.-S.; Xia, Z.-G.; Zhou, S.-S. The progress of national malaria elimination and epidemiological characteristics of malaria in China in 2017. Chin. J. Parasitol. Parasit. Dis. 2018, 36, 201–209. [Google Scholar]
- Zhang, L.; Feng, J.; Zhang, S.-S.; Xia, Z.-G.; Zhou, S.-S. Epidemiological characteristics of malaria and the progress towards its elimination in China in 2018. Chin. J. Parasitol. Parasit. Dis. 2019, 37, 241–247. [Google Scholar]
- Zhang, L.; Feng, J.; Xia, Z.-G.; Zhou, S.-S. Epidemiological characteristics of malaria and progress on its elimination in China in 2019. Chin. J. Parasitol. Parasit. Dis. 2020, 38, 133–138. [Google Scholar]
- Zhang, L.; Feng, J.; Tu, H.; Yin, J.-H.; Xia, Z.-G. Malaria epidemiology in China in 2020. Chin. J. Parasitol. Parasit. Dis. 2021, 39, 195–199. [Google Scholar]
- Feng, J.; Zhang, L.; Xia, Z.-G.; Xiao, N. Malaria elimination in China: An eminent milestone in the anti-malaria campaign and challenges in the post-elimination stage. Chin. J. Parasitol. Parasit. Dis. 2021, 39, 421–428. [Google Scholar]
- Zhou, S.; Li, Z.; Cotter, C.; Zheng, C.; Zhang, Q.; Li, H.; Zhou, S.; Zhou, X.; Yu, H.; Yang, W. Trends of imported malaria in China 2010–2014: Analysis of surveillance data. Malar. J. 2016, 15, 39. [Google Scholar] [CrossRef]
- Li, Z.; Yang, Y.; Xiao, N.; Zhou, S.; Lin, K.; Wang, D.; Zhang, Q.; Jiang, W.; Li, M.; Feng, X.; et al. Malaria imported from Ghana by returning gold miners, China, 2013. Emerg. Infect. Dis. 2015, 21, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Xu, H.F.; Zhu, X.Y. Epidemiological status of malaria in the world and imported malaria in China. Chin. J. Front. Health Quar. 2013, 6, 425–427. [Google Scholar]
- Yang, W.Z.; Zhou, X.N. New challenges of malaria elimination in China. Chin. J. Prev. Med. 2016, 50, 289–291. [Google Scholar]
- Cao, J.; Liu, Y.; Cao, Y.; Zhu, G.; Zhou, S. Sustained Challenge to Malaria Elimination in China: Imported Malaria. Chin. J. Parasitol. Parasit. Dis. 2018, 36, 93–96. [Google Scholar]
- Feng, J.; Li, J.; Yan, H.; Feng, X.; Xia, Z. Evaluation of antimalarial resistance marker polymorphism in returned migrant workers in China. Antimicrob. Agents Chemother. 2015, 59, 326–330. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Juma, D.W.; Omondi, A.A.; Ingasia, L.; Opot, B.; Cheruiyot, A.; Yeda, R.; Okudo, C.; Cheruiyot, J.; Muiruri, P.; Ngalah, B.; et al. Trends in drug resistance codons in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes in Kenyan parasites from 2008 to 2012. Malar. J. 2014, 13, 250. [Google Scholar] [CrossRef] [PubMed][Green Version]
- WHO. Report of the Technical Consultation on Intermittent Preventive Treatment in Infants (IPTi), Technical Expert Group on Preventive Chemotherapy; World Health Organization: Geneva, Switzerland, 2009. [Google Scholar]
- Alam, M.T.; De Souza, D.K.; Vinayak, S.; Griffing, S.M.; Poe, A.C.; Duah, N.O.; Ghansah, A.; Asamoa, K.; Slutsker, L.; Wilson, M.D.; et al. Selective sweeps and genetic lineages of Plasmodium falciparum drug-resistant alleles in Ghana. J. Infect. Dis. 2011, 203, 220–227. [Google Scholar] [CrossRef]
- Grais, R.F.; Laminou, I.M.; Woi-Messe, L.; Makarimi, R.; Bouriema, S.H.; Langendorf, C.; Amambua-Ngwa, A.; D’Alessandro, U.; Guerin, P.J.; Fandeur, T.; et al. Molecular markers of resistance to amodiaquine plus sulfadoxine-pyrimethamine in an area with seasonal malaria chemoprevention in south central Niger. Malar. J. 2018, 17, 98. [Google Scholar] [CrossRef]
- Duah, N.O.; Quashie, N.B.; Abuaku, B.K.; Sebeny, P.J.; Kronmann, K.C.; Koram, K.A. Surveillance of molecular markers of Plasmodium falciparum resistance to sulphadoxine-pyrimethamine 5 years after the change of malaria treatment policy in Ghana. Am. J. Trop. Med. Hyg. 2012, 87, 996–1003. [Google Scholar] [CrossRef]
- Vinayak, S.; Alam, T.; Mixson-Hayden, T.; McCollum, A.M.; Sem, R.; Shah, N.K.; Lim, P.; Muth, S.; Rogers, W.O.; Fandeur, T.; et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog. 2010, 6, e1000830. [Google Scholar] [CrossRef]
- Amenga-Etego, L.N.; Asoala, V.; Agongo, G.; Jacob, C.; Goncalves, S.; Awandare, G.A.; Rockett, K.A.; Kwiatkowski, D. Temporal evolution of sulfadoxine-pyrimethamine resistance genotypes and genetic diversity in response to a decade of increased interventions against Plasmodium falciparum in northern Ghana. Malar. J. 2021, 20, 152. [Google Scholar] [CrossRef] [PubMed]
- Nkoli Mandoko, P.; Rouvier, F.; Matendo Kakina, L.; Moke Mbongi, D.; Latour, C.; Losimba Likwela, J.; Ngoyi Mumba, D.; Bi Shamamba, S.K.; Tamfum Muyembe, J.-J.; Muepu Tshilolo, L.; et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine/pyrimethamine in the Democratic Republic of the Congo: Emergence of highly resistant pfdhfr/pfdhps alleles. J. Antimicrob. Chemother. 2018, 73, 2704–2715. [Google Scholar] [CrossRef] [PubMed]
- Divala, T.H.; Cohee, L.M.; Laufer, M.K. The remarkable tenacity of sulfadoxine-pyrimethamine. Lancet Infect. Dis. 2019, 19, 460–461. [Google Scholar] [CrossRef] [PubMed]
- Al-Saai, S.; Kheir, A.; Abdel-Muhsin, A.-M.A.; Al-Ghazali, A.; Nwakanma, D.; Swedberg, G.; Babiker, H.A. Distinct haplotypes of dhfr and dhps among Plasmodium falciparum isolates in an area of high level of sulfadoxine-pyrimethamine (SP) resistance in eastern Sudan. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2009, 9, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Pearce, R.J.; Pota, H.; Evehe, M.-S.B.; Ba, E.-H.; Mombo-Ngoma, G.; Malisa, A.L.; Ord, R.; Inojosa, W.; Matondo, A.; Diallo, D.A.; et al. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009, 6, e1000055. [Google Scholar] [CrossRef] [PubMed]
- McCollum, A.M.; Poe, A.C.; Hamel, M.; Huber, C.; Zhou, Z.; Shi, Y.P.; Ouma, P.; Vulule, J.; Bloland, P.; Slutsker, L.; et al. Antifolate resistance in Plasmodium falciparum: Multiple origins and identification of novel dhfr alleles. J. Infect. Dis. 2006, 194, 189–197. [Google Scholar] [CrossRef][Green Version]
- Koukouikila-Koussounda, F.; Bakoua, D.; Fesser, A.; Nkombo, M.; Vouvoungui, C.; Ntoumi, F. High prevalence of sulphadox-ine-pyrimethamine resistance-associated mutations in Plasmodium falciparum field isolates from pregnant women in Brazzaville, Republic of Congo. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 33, 32–36. [Google Scholar]
- Frosch, A.E.P.; Laufer, M.K.; Mathanga, D.P.; Takala-Harrison, S.; Skarbinski, J.; Claassen, C.W.; Dzinjalamala, F.K.; Plowe, C.V. Return of widespread chloro-quine-sensitive Plasmodium falciparum to Malawi. J. Infect. Dis. 2014, 210, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, M.; Alifrangis, M.; Roper, C.; Plowe, C.V. Monitoring antifolate resistance in intermittent preventive therapy for malaria. Trends Parasitol. 2013, 29, 497–504. [Google Scholar] [CrossRef]
- van Lenthe, M.; van der Meulen, R.; Lassovski, M.; Ouabo, A.; Bakula, E.; Badio, C.; Cibenda, D.; Okell, L.; Piriou, E.; Grignard, L.; et al. Markers of sulfadoxine-pyrimethamine resistance in Eastern Democratic Republic of Congo; implications for malaria chemoprevention. Malar. J. 2019, 18, 430. [Google Scholar] [CrossRef]
- Naidoo, I.; Roper, C. Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol. 2013, 29, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Stokes, B.H.; Dhingra, S.K.; Rubiano, K.; Mok, S.; Straimer, J.; Gnadig, N.F.; Deni, I.; Schindler, K.A.; Bath, J.R.; Ward, K.E.; et al. Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. eLife 2021, 10, e66277. [Google Scholar] [CrossRef] [PubMed]
| dhfr | First round | 5′-TCCTTTTTATGATGGAACAAG-3′ |
| 5′-AGTATATACATCGCTAACAGA-3′ | ||
| Second round | 5′-TTTATGATGGAACAAGTCTGC-3′ | |
| 5′-ACTCATTTTCATTTATTTCTGG-3′ | ||
| dhps | First round | 5′-AACCTAAACGTGCTGTTCAA-3′ |
| 5′-AATTGTGTGATTTGTCCACAA-3′ | ||
| Second round | 5′-ATGATAAATGAAGGTGCTAG-3′ | |
| 5′-TCATTTTGTTGTTCATCATGT-3′ |
| Gene | Year | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duah’s Study | Our Study | Amenga-Etego’s Study | ||||||||||||||
| 2003– 2004 | 2005– 2006 | 2007– 2008 | 2010 | 2013 | 2009 | 2010 | 2011 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | |||
| dhfr | Point allele | I51 | 49 | 49.1 | 60.8 | 60.7 | 83.2 | - | - | - | - | - | - | - | - | - |
| R59 | 52.2 | 61.9 | 71.9 | 80.9 | 94.9 | - | - | - | - | - | - | - | - | - | ||
| N108 | 69.7 | 68.9 | 76.6 | 80.9 | 96.4 | - | - | - | - | - | - | - | - | - | ||
| Haplotype | IN | 9 | 11.1 | 7.6 | 3.4 | 1.5 | 3.3 | 2.2 | 1.8 | 6.1 | 1.2 | 1.1 | 5.7 | 4.8 | 2.2 | |
| RN | 15 | 22 | 17.3 | 21.2 | 13.1 | 7.5 | 9.2 | 14.5 | 6.1 | 20.7 | 18.5 | 7.1 | 16.1 | 24.6 | ||
| IRN | 35.3 | 28.8 | 46.1 | 53.9 | 81.8 | 80 | 77.2 | 76.4 | 66.7 | 67.5 | 73.6 | 79.8 | 70.7 | 67.9 | ||
| dhps | Point allele | G437 | 59.2 | 71.5 | 80 | 77.6 | 95.2 | 2.5 | 33.7 | 9.1 | 39.4 | 55.4 | 64 | 71.2 | 69.5 | 78.2 |
| E540 | 0 | 0.5 | 0.2 | 1.1 | 3.4 | 0 | 0 | 0 | 3.0 | 0 | 1.7 | 1.1 | 1.3 | 0 | ||
| S613 | - | - | - | - | 2.7 | 14.2 | 19 | 18.2 | 6.1 | 7.1 | 3.4 | 3.4 | 4.1 | 6.9 | ||
| Haplotype | G437E540 | 0 | 0.5 | 0.2 | 1.1 | 3.4 | 0 | 0 | 0 | 3.0 | 0 | 1.7 | 1.1 | 1.3 | 0 | |
| A436G437S613 | - | - | - | - | 1.4 | 2.5 | 3.8 | 3.6 | 6.1 | 2.5 | 9.6 | 4.6 | 8.0 | 4.2 | ||
| dhfr/dhps haplotype combination | Triple 1 | - | - | - | - | 7.5 | 3.5 | 0 | 0 | 0 | 5.5 | 1.4 | 1.3 | 0 | 2.8 | |
| Quadruple 2 | - | - | - | - | 50.7 | 43.9 | 49.4 | 87.5 | 59.1 | 45.2 | 25.7 | 10.1 | 21.1 | 20.6 | ||
| Quintuple 3 | - | - | - | - | 30.1 | 52.6 | 50.6 | 12.5 | 40.9 | 49.3 | 72.9 | 88.6 | 78.9 | 76.6 | ||
| Sextuple 4 | - | - | - | - | 1.4 | - | - | - | - | - | - | - | - | - | ||
| dhfr/dhps Haplotype Combinations | Prevalence % (n) | |
|---|---|---|
| Triple | R59N108/G437 | 7.5 (11/146) |
| Quadruple | I51R59N108/G437 | 41.1 (60/146) |
| I51R59N108/A436 | 3.4 (5/146) | |
| R59N108/A436G437 | 3.4 (5/146) | |
| R59N108/G437E540 | 0.7 (1/146) | |
| R59N108/G437S613 | 0.7 (1/146) | |
| I51N108/A436G437 | 0.7 (1/146) | |
| I51N108/G437E540 | 0.7 (1/146) | |
| Quintuple | I51R59N108/A436G437 | 27.4 (40/146) |
| I51R59N108/G437E540 | 2.1 (3/146) | |
| I51R59N108/Y436S613 | 0.7 (1/146) | |
| Sextuple | I51R59N108/A436G437S613 | 1.4 (2/146) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quan, H.; Yu, P.; Kassegne, K.; Shen, H.-M.; Chen, S.-B.; Chen, J.-H. Polymorphism of Drug Resistance Genes dhfr and dhps in Plasmodium falciparum Isolates among Chinese Migrant Workers Who Returned from Ghana in 2013. Trop. Med. Infect. Dis. 2023, 8, 504. https://doi.org/10.3390/tropicalmed8110504
Quan H, Yu P, Kassegne K, Shen H-M, Chen S-B, Chen J-H. Polymorphism of Drug Resistance Genes dhfr and dhps in Plasmodium falciparum Isolates among Chinese Migrant Workers Who Returned from Ghana in 2013. Tropical Medicine and Infectious Disease. 2023; 8(11):504. https://doi.org/10.3390/tropicalmed8110504
Chicago/Turabian StyleQuan, Hong, Peng Yu, Kokouvi Kassegne, Hai-Mo Shen, Shen-Bo Chen, and Jun-Hu Chen. 2023. "Polymorphism of Drug Resistance Genes dhfr and dhps in Plasmodium falciparum Isolates among Chinese Migrant Workers Who Returned from Ghana in 2013" Tropical Medicine and Infectious Disease 8, no. 11: 504. https://doi.org/10.3390/tropicalmed8110504
APA StyleQuan, H., Yu, P., Kassegne, K., Shen, H.-M., Chen, S.-B., & Chen, J.-H. (2023). Polymorphism of Drug Resistance Genes dhfr and dhps in Plasmodium falciparum Isolates among Chinese Migrant Workers Who Returned from Ghana in 2013. Tropical Medicine and Infectious Disease, 8(11), 504. https://doi.org/10.3390/tropicalmed8110504









