Single Nucleotide Variants in the TLR1, TLR2 and TLR6 Genes: A Case–Control Study in a Colombian Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Declaration
2.2. Study Population
2.3. Study Design
2.4. SNVs Selection
2.5. DNA Extraction
2.6. Genotyping
2.7. Sequencing
2.8. Statistical Analysis
3. Results
3.1. Genotype and Allelic Distribution of SNVs
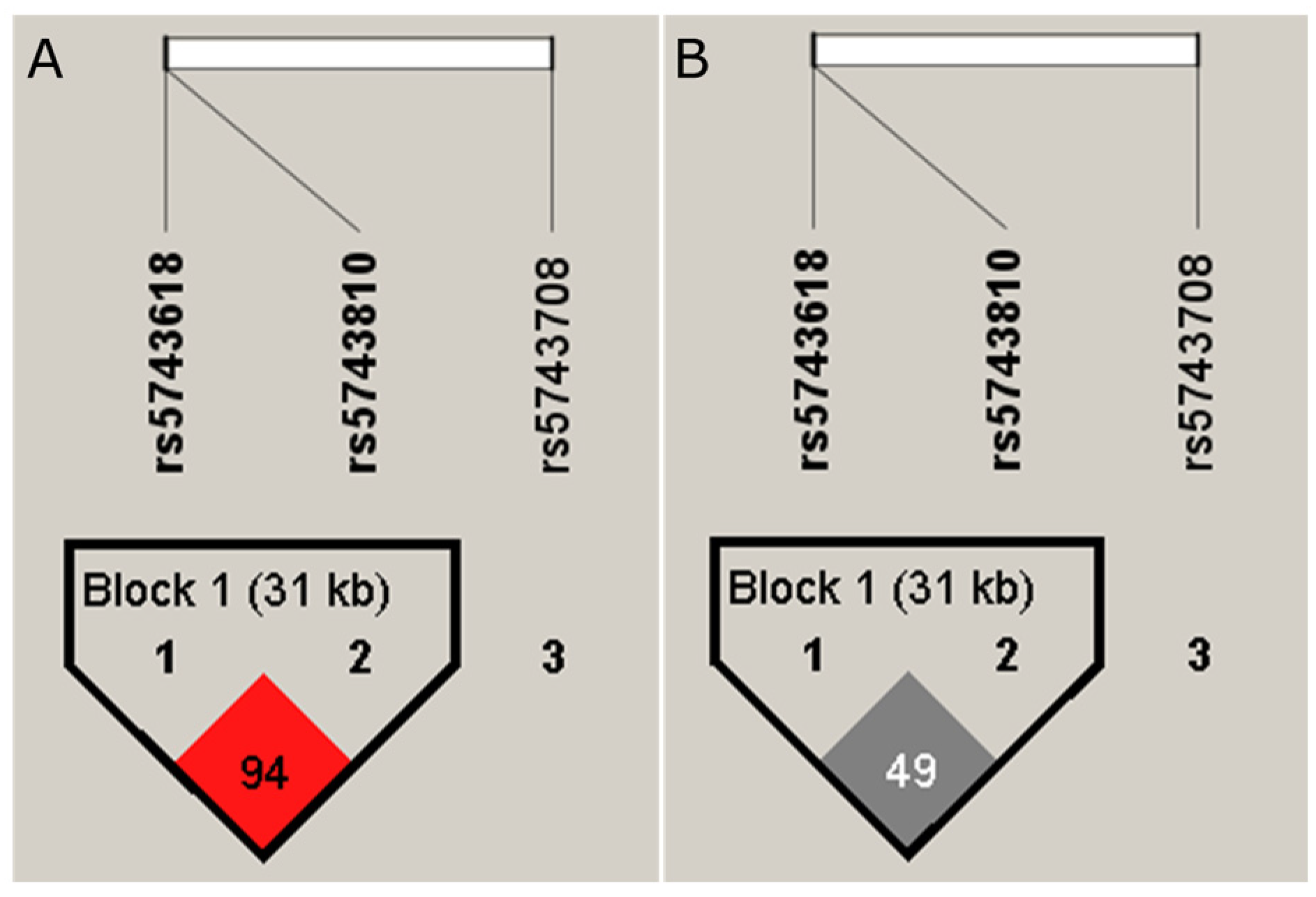
3.2. Haplotype Analysis
4. Discussion
5. Conclusions
6. Study Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alemu Belachew, W.; Naafs, B. Position statement: LEPROSY: Diagnosis, treatment, and follow-up. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, R.O.; Schmitz, V.; Silva, B.J.d.A.; Dias, A.A.; de Souza, B.J.; Barbosa, M.G.d.M.; Esquenazi, D.d.A.; Pessolani, M.C.V.; Sarno, E.N. Innate Immune Responses in Leprosy. Front. Immunol. 2018, 9, 518. [Google Scholar] [CrossRef] [PubMed]
- Sartori, P.V.U.; Penna, G.O.; Bührer-Sékula, S.; Pontes, M.A.A.; Gonçalves, H.S.; Cruz, R.; Virmond, M.C.L.; Dias-Baptista, I.M.F.; Rosa, P.S.; Penna, M.L.F.; et al. Human Genetic Susceptibility of Leprosy Recurrence. Sci. Rep. 2020, 10, 1284. [Google Scholar] [CrossRef] [PubMed]
- Chavarro-Portillo, B.; Soto, C.Y.; Guerrero, M.I. Mycobacterium leprae’s evolution and environmental adaptation. Acta Tropica. 2019, 197, 105041. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Leprosy (Hansen’s Disease). Available online: https://www.who.int/news-room/fact-sheets/detail/leprosy (accessed on 29 September 2021).
- Lastória, J.C.; de Abreu, M.A.M.M. Leprosy: Review of the epidemiological, clinical, and etiopathogenic aspects—Part 1. An. Bras. Dermatol. 2014, 89, 205–218. [Google Scholar] [CrossRef]
- Kamath, S.; Vaccaro, S.A.; Rea, T.H.; Ochoa, M.T. Recognizing and managing the immunologic reactions in leprosy. J. Am. Acad. Dermatol. 2014, 71, 795–803. [Google Scholar] [CrossRef]
- Fava, V.; Orlova, M.; Cobat, A.; Alcaïs, A.; Mira, M.; Schurr, E. Genetics of leprosy reactions: An overview. Mem. Inst. Oswaldo Cruz. 2012, 107, 132–142. [Google Scholar] [CrossRef]
- Pinheiro, R.O.; de Souza Salles, J.; Sarno, E.N.; Sampaio, E.P. Mycobacterium leprae –host-cell interactions and genetic determinants in leprosy: An overview. Future Microbiol. 2011, 6, 217–230. [Google Scholar] [CrossRef]
- Ochoa, M.T. Aspectos inmunológicos. In La lepra: Una Enfermedad Vgente; Guerrero, M.I., Hernández, C.A., Rodríguez, G., Eds.; Centro Dermatológico Federico Lleras Acosta: Bogotá, Colombia, 2019; Panamericana Formas e Impresos; pp. 249–262. ISBN 978-958-59331-2-5. [Google Scholar]
- Gutiérrez, L.D.; Tovar-Parra, D. Genética de la susceptibilidad a Mycobacterium leprae. In La lepra: Una Enfermedad Vgente; Guerrero, M.I., Hernández, C.A., Rodríguez, G., Eds.; Centro Dermatológico Federico Lleras Acosta: Bogotá, Colombia, 2020; Panamericana Formas e Impresos; pp. 263–280. ISBN 978-958-59331-2-5. [Google Scholar]
- Takeda, K.; Akira, S. Toll-Like Receptors. Curr. Protoc. Immunol. 2015, 109, 14.12.1–14.12.10. [Google Scholar] [CrossRef]
- Bochud, P.; Hawn, T.R.; Siddiqui, M.R.; Saunderson, P.; Britton, S.; Abraham, I.; Argaw, A.T.; Janer, M.; Zhao, L.P.; Kaplan, G.; et al. Toll-Like Receptor 2 (TLR2) Polymorphisms Are Associated with Reversal Reaction in Leprosy. J. Infect. Dis. 2008, 197, 253–261. [Google Scholar] [CrossRef]
- Shey, M.S.; Randhawa, A.K.; Bowmaker, M.; Smith, E.; Scriba, T.J.; de Kock, M.; Mahomed, H.; Hussey, G.; Hawn, T.R.; Hanekom, W.A. Single nucleotide polymorphisms in toll-like receptor 6 are associated with altered lipopeptide- and mycobacteria-induced interleukin-6 secretion. Genes. Immun. 2010, 11, 561–572. [Google Scholar] [CrossRef]
- Mattos, K.A.; Oliveira, V.G.C.; D’avila, H.; Rodrigues, L.S.; Pinheiro, R.O.; Sarno, E.N.; Pessolani, M.C.V.; Bozza, P.T. TLR6-Driven Lipid Droplets in Mycobacterium leprae- Infected Schwann Cells: Immunoinflammatory Platforms Associated with Bacterial Persistence. J. Immunol. 2011, 187, 2548–2558. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Gochhait, S.; Malhotra, D.; Pettersson, F.H.; Teo, Y.Y.; Khor, C.C.; Rautanen, A.; Chapman, S.J.; Mills, T.C.; Srivastava, A.; et al. Leprosy and the Adaptation of Human Toll-Like Receptor 1. PLoS Pathog. 2010, 6, e1000979. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Lyle, E.A.; Omueti, K.O.; Stepensky, V.A.; Yegin, O.; Alpsoy, E.; Hamann, L.; Schumann, R.R.; Tapping, R.I. Cutting Edge: A Common Polymorphism Impairs Cell Surface Trafficking and Functional Responses of TLR1 but Protects against Leprosy. J. Immunol. 2007, 178, 7520–7524. [Google Scholar] [CrossRef]
- Liu, H.; Bao, F.; Irwanto, A.; Fu, X.; Lu, N.; Yu, G.; Yu, Y.; Sun, Y.; Low, H.; Li, Y.; et al. An association study of TOLL and CARD with leprosy susceptibility in Chinese population. Hum. Mol. Genet. 2013, 22, 4430–4437. [Google Scholar] [CrossRef] [PubMed]
- The rs5743708 Gene Polymorphism in the TLR2 Gene Contributes to the Risk of Tuberculosis Disease—PMC. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4637765/ (accessed on 15 October 2022).
- Schurz, H.; Daya, M.; Möller, M.; Hoal, E.G.; Salie, M. TLR1, 2, 4, 6 and 9 Variants Associated with Tuberculosis Susceptibility: A Systematic Review and Meta-Analysis. PLoS ONE 2015, 10, e0139711. [Google Scholar] [CrossRef] [PubMed]
- Maciel-Fiuza, M.F.; Costa, P.d.S.S.; Kowalski, T.W.; Schuler-Faccini, L.; Bonamigo, R.R.; Vetoratto, R.; Eidt, L.M.; de Moraes, P.C.; Silveira, M.I.d.S.; Camargo, L.M.A.; et al. Evaluation of Polymorphisms in Toll-Like Receptor Genes as Biomarkers of the Response to Treatment of Erythema Nodosum Leprosum. Front. Med. 2022, 8, 713143. [Google Scholar] [CrossRef] [PubMed]
- Little, J.; Higgins, J.P.T.; Ioannidis, J.P.A.; Moher, D.; Gagnon, F.; von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. STrengthening the REporting of Genetic Association Studies (STREGA)—An Extension of the STROBE Statement. PLOS Med. 2009, 6, e1000022. [Google Scholar] [CrossRef]
- Kelsey, J.L.; Whittemore, A.S.; Evans, A.S.; Thompson, W.D. Methods in Observational Epidemiology; Oxford University Press: Oxford, UK, 1996. [Google Scholar]
- Hawn, T.R.; Misch, E.A.; Dunstan, S.J.; Thwaites, G.E.; Lan, N.T.N.; Quy, H.T.; Chau, T.T.H.; Rodrigues, S.; Nachman, A.; Janer, M.; et al. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur. J. Immunol. 2007, 37, 2280–2289. [Google Scholar] [CrossRef]
- Sole, X.; Guino, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Krutzik, S.R.; Ochoa, M.T.; Sieling, P.A.; Uematsu, S.; Ng, Y.W.; Legaspi, A.; Liu, P.T.; Cole, S.T.; Godowski, P.J.; Maeda, Y.; et al. Activation and regulation of toll-like receptors 2 and 1 in human leprosy. Nat. Med. 2003, 9, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.S.; Kim, S.E.; Heo, J.Y.; Lee, M.E.; Kim, H.M.; Paik, S.-G.; Lee, H.; Lee, J.-O. Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Hart, B.E.; Tapping, R.I. Genetic Diversity of Toll-Like Receptors and Immunity to M. leprae Infection. J. Trop. Med. 2012, 2012, 415057. [Google Scholar] [CrossRef]
- National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/snp/rs5743618?horizontal_tab=true#clinical_significance (accessed on 11 January 2022).
- Ossa, H.; Aquino, J.; Pereira, R.; Ibarra, A.; Ossa, R.H.; Pérez, L.A.; Granda, J.D.; Lattig, M.C.; Groot, H.; de Carvalho, E.F.; et al. Outlining the Ancestry Landscape of Colombian Admixed Populations. PLoS ONE 2016, 11, e0164414. [Google Scholar] [CrossRef]
- Hart, B.E.; Tapping, R.I. Differential Trafficking of TLR1 I602S Underlies Host Protection against Pathogenic Mycobacteria. J. Immunol. 2012, 189, 5347–5355. [Google Scholar] [CrossRef]
- Thurow, H.S.; Sarturi, C.R.; Fallavena, P.R.V.; Paludo, F.J.d.O.; Picanço, J.B.; Fraga, L.R.; Graebin, P.; de Souza, V.C.; Dias, F.S.; Nóbrega, O.d.T.; et al. Very Low Frequencies of Toll-Like Receptor 2 Supposed-2029T and 2258A (RS5743708) Mutant Alleles in Southern Brazilian Critically Ill Patients: ¿Would It Be a Lack of Worldwide-Accepted Clinical Applications of Toll-Like Receptor 2 Variants? Genet. Test. Mol. Biomark. 2010, 14, 405–419. [Google Scholar] [CrossRef]
- Ogus, A.; Yoldas, B.; Ozdemir, T.; Uguz, A.; Olcen, S.; Keser, I.; Coskun, M.; Cilli, A.; Yegin, O. The Arg753Gln polymorphism of the human Toll-like receptor 2 gene in tuberculosis disease. Eur. Respir. J. 2004, 23, 219–223. [Google Scholar] [CrossRef]
- National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/snp/rs5743708?horizontal_tab=true (accessed on 11 January 2022).
- Hu, L.; Tao, H.; Tao, X.; Tang, X.; Xu, C. TLR2 Arg753Gln Gene Polymorphism Associated with Tuberculosis Susceptibility: An Updated Meta-Analysis. BioMed Res. Int. 2019, 2019, 2628101. [Google Scholar] [CrossRef]
- Bochud, P.-Y.; Hawn, T.R.; Aderem, A. Cutting Edge: A Toll-Like Receptor 2 Polymorphism That Is Associated with Lepromatous Leprosy Is Unable to Mediate Mycobacterial Signaling. J. Immunol. 2003, 170, 3451–3454. [Google Scholar] [CrossRef]
- National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/snp/rs5743810?horizontal_tab=true (accessed on 11 January 2022).
- Stappers, M.H.T.; Thys, Y.; Oosting, M.; Plantinga, T.S.; Ioana, M.; Reimnitz, P.; Mouton, J.W.; Netea, M.G.; Joosten, L.A.B.; Gyssens, I.C. TLR1, TLR2, and TLR6 Gene Polymorphisms Are Associated with Increased Susceptibility to Complicated Skin and Skin Structure Infections. J. Infect. Dis. 2014, 210, 311–318. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schoffelen, T.; Ammerdorffer, A.; Hagenaars, J.C.J.P.; Bleeker-Rovers, C.P.; Wegdam-Blans, M.C.; Wever, P.C.; Joosten, L.A.B.; van der Meer, J.W.M.; Sprong, T.; Netea, M.G.; et al. Genetic Variation in Pattern Recognition Receptors and Adaptor Proteins Associated with Development of Chronic Q Fever. J. Infect. Dis. 2015, 212, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Varzari, A.; Deyneko, I.V.; Tudor, E.; Grallert, H.; Illig, T. Synergistic effect of genetic polymorphisms in TLR6 and TLR10 genes on the risk of pulmonary tuberculosis in a Moldavian population. Innate Immun. 2021, 27, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Instituto Nacional de Salud. Lepra (Enfermedad de Hansen). Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2020_Boletin_epidemiologico_semana_3.pdf (accessed on 29 September 2021).
| Gen/SNP | Allele/ Genotype | Cases | Controls | χ2 | p-Value | ||
|---|---|---|---|---|---|---|---|
| (n = 114) | (n = 456) | ||||||
| N | Frequency | n | Frequency | ||||
| TLR1 (rs5743618) | A | 166 | 0.73 | 647 | 0.71 | 0.309 | 0.5778 |
| C | 62 | 0.27 | 265 | 0.29 | |||
| A/A | 61 | 0.54 | 231 | 0.51 | 0.314 | 0.8543 * | |
| A/C | 44 | 0.39 | 185 | 0.41 | |||
| C/C | 9 | 0.08 | 40 | 0.09 | |||
| TLR2 (rs5743708) | G | 227 | 1 | 909 | 1 | 0.062 | 0.8022 * |
| A | 1 | 0 | 3 | 0 | |||
| G/A | 1 | 0.01 | 3 | 0.01 | 0.062 | 0.8019 | |
| G/G | 113 | 0.99 | 453 | 0.99 | |||
| TLR6 (rs5743810) | G | 194 | 0.85 | 738 | 0.81 | 2.12 | 0.1451 |
| A | 34 | 0.15 | 174 | 0.19 | |||
| A/A | 3 | 0.03 | 14 | 0.03 | 2.57 | 0.2760 | |
| G/A | 28 | 0.25 | 146 | 0.32 | |||
| G/G | 83 | 0.73 | 296 | 0.65 | |||
| Gene/SNV | Model | Genotype | Cases | Controls | OR (95% CI) | p-Value * |
|---|---|---|---|---|---|---|
| n (Frequency) | n (Frequency) | |||||
| TLR1 rs5743618 | Codominant | A/A | 61 | 231 (50.7%) | 1 | 0.99 |
| −53.50% | ||||||
| C/A | 44 | 185 (40.6%) | 0.98 (0.60–1.59) | |||
| −38.60% | ||||||
| C/C | 9 | 40 | 0.95 (0.40–2.25) | |||
| −7.90% | −8.80% | |||||
| Dominant | A/A | 61 (53.5%) | 231 (50.7%) | 1 | 0.91 | |
| C/A-C/C | 53 | 225 (49.3%) | 0.97 (0.61–1.54) | |||
| −46.50% | ||||||
| Recessive | A/A-C/A | 105 (92.1%) | 416 (91.2%) | 1 | 0.92 | |
| C/C | 9 | 40 (8.8%) | 0.96 (0.41–2.21) | |||
| −7.90% | ||||||
| Over-dominant | A/A-C/C | 70 (61.4%) | 271 (59.4%) | 1 | 0.95 | |
| C/A | 44 (38.6%) | 185 (40.6%) | 0.99 (0.62–1.58) | |||
| Log-additive | --- | --- | --- | 0.98 (0.68–1.40) | 0.89 | |
| TLR6 rs5743810 | Codominant | G/G | 83 (72.8%) | 296 (64.9%) | 1 | 0.18 |
| A/G | 28 (24.6%) | 146 (32%) | 0.61 (0.6–1.04) | |||
| A/A | 3 (2.6%) | 14 (3.1%) | 0.75 (0.18–3.04) | |||
| Dominant | G/G | 83 (72.8%) | 296 (64.9%) | 1 | 0.066 | |
| A/G-A/A | 31 (27.2%) | 160 (35.1%) | 0.63 (0.38–1.04) | |||
| Recessive | G/G-A/G | 111 (97.4%) | 442 (96.9%) | 1 | 0.83 | |
| A/A | 3 (2.6%) | 14 (3.1%) | 0.86 (0.21–3.47) | |||
| Over-dominant | G/G-A/A | 86 (75.4%) | 310 (68%) | 1 | 0.07 | |
| A/G | 28 (24.6%) | 146 (32%) | 0.62 (0.37–1.05) | |||
| Log-additive | --- | --- | 1.46 (0.93–2.29) | 0.68 (0.44–1.08) | 0.092 |
| Gene/SNV | Model | Genotype | Cases n (Frequency) | Controls n (Frequency) | OR (95% CI) | p-Value * |
|---|---|---|---|---|---|---|
| TLR1 rs5743618 | Codominant | A/A | 32 (55.2%) | 107 (46.7%) | 1 | 0.46 |
| C/A | 22 (37.9%) | 100 (43.7%) | 0.68 (0.33–1.41) | |||
| C/C | 4 (6.9%) | 22 (9.6%) | 0.54 (0.14–2.06) | |||
| Dominant | A/A | 32 (55.2%) | 107 (46.7%) | 1 | 0.23 | |
| C/A-C/C | 26 (44.8%) | 122 (53.3%) | 0.66 (0.33–1.31) | |||
| Recessive | A/A-C/A | 54 (93.1%) | 207 (90.4%) | 1 | 0.5 | |
| C/C | 4 (6.9%) | 22 (9.6%) | 0.65 (0.18–2.35) | |||
| Over-dominant | A/A-C/C | 36 (62.1%) | 129 (56.3%) | 1 | 0.4 | |
| C/A | 22 (37.9%) | 100 (43.7%) | 0.74 (0.37–1.50) | |||
| Log-additive | --- | --- | --- | 0.71 (0.41–1.23) | 0.22 | |
| TLR6 rs5743810 | Codominant | G/G | 45 (77.6%) | 147 (64.2%) | 1 | 0.049 |
| A/G | 11 (19%) | 74 (32.3%) | 0.37 (0.16–0.86) | |||
| A/A | 2 (3.5%) | 8 (3.5%) | 0.56 (0.09–3.52) | |||
| Dominant | G/G | 45 (77.6%) | 147 (64.2%) | 1 | 0.016 | |
| A/G-A/A | 13 (22.4%) | 82 (35.8%) | 0.39 (0.17–0.87) | |||
| Recessive | G/G-A/G | 56 (96.5%) | 221 (96.5%) | 1 | 0.74 | |
| A/A | 2 (3.5%) | 8 (3.5%) | 0.74 (0.12–4.53) | |||
| Over-dominant | G/G-A/A | 47 (81%) | 155 (67.7%) | 1 | 0.019 | |
| A/G | 11 (19%) | 74 (32.3%) | 0.38 (0.16–0.88) | |||
| Log-additive | --- | --- | --- | 0.49 (0.25–0.97) | 0.032 |
| Haplotype | rs5743618 | rs5743708 | rs5743810 | Frequency | OR (95% CI) | p-Value * |
|---|---|---|---|---|---|---|
| 1 | A | G | G | 0.7044 | 1 | --- |
| 2 | C | G | A | 0.1756 | 0.65 (0.40–1.06) | 0.082 |
| 3 | C | G | G | 0.1096 | 1.86 (1.11–3.10) | 0.019 |
| Female Subgroup, n = 283. | ||||||
|---|---|---|---|---|---|---|
| Haplotype | rs5743618 | rs5743708 | rs5743810 | Frequency | OR (95% CI) | p-Value * |
| 1 | A | G | G | 0.71 | 1 | --- |
| 2 | C | G | A | 0.17 | 0.81 (0.42–1.59) | 0.55 |
| 3 | C | G | G | 0.10 | 2.39 (1.21–4.72) | 0.013 |
| 4 | A | rs5743708 G | A | 0.01 | 6.92 (1.08–44.25) | 0.042 |
| Global haplotype association p-value: 0.017 | ||||||
| Male Subgroup, n = 287. | ||||||
| Haplotype | rs5743618 | rs5743708 | rs5743810 | Frequency | OR (95% CI) | p-value * |
| 1 | A | G | G | 0.6928 | 1 | --- |
| 2 | C | G | A | 0.181 | 0.51 (0.25–1.01) | 0.056 |
| 3 | C | G | G | 0.1207 | 1.32 (0.59–2.93) | 0.5 |
| Global haplotype association p-value: 0.15 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutierrez-Castañeda, L.D.; Acosta, C.R.; Bustos, M.A.; García, D.K.; Bohada, D.P.; Rodríguez, R.; Guerrero, M.I. Single Nucleotide Variants in the TLR1, TLR2 and TLR6 Genes: A Case–Control Study in a Colombian Population. Trop. Med. Infect. Dis. 2023, 8, 473. https://doi.org/10.3390/tropicalmed8100473
Gutierrez-Castañeda LD, Acosta CR, Bustos MA, García DK, Bohada DP, Rodríguez R, Guerrero MI. Single Nucleotide Variants in the TLR1, TLR2 and TLR6 Genes: A Case–Control Study in a Colombian Population. Tropical Medicine and Infectious Disease. 2023; 8(10):473. https://doi.org/10.3390/tropicalmed8100473
Chicago/Turabian StyleGutierrez-Castañeda, Luz D., Carmen R. Acosta, Mónica A. Bustos, Diana K. García, Diana P. Bohada, Raúl Rodríguez, and Martha Inirida Guerrero. 2023. "Single Nucleotide Variants in the TLR1, TLR2 and TLR6 Genes: A Case–Control Study in a Colombian Population" Tropical Medicine and Infectious Disease 8, no. 10: 473. https://doi.org/10.3390/tropicalmed8100473
APA StyleGutierrez-Castañeda, L. D., Acosta, C. R., Bustos, M. A., García, D. K., Bohada, D. P., Rodríguez, R., & Guerrero, M. I. (2023). Single Nucleotide Variants in the TLR1, TLR2 and TLR6 Genes: A Case–Control Study in a Colombian Population. Tropical Medicine and Infectious Disease, 8(10), 473. https://doi.org/10.3390/tropicalmed8100473






