Co-Radiation of Leptospira and Tenrecidae (Afrotheria) on Madagascar
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Sample
2.2. Leptospira Detection and Sequencing
2.3. Statistical Analyses
2.4. Phylogeny
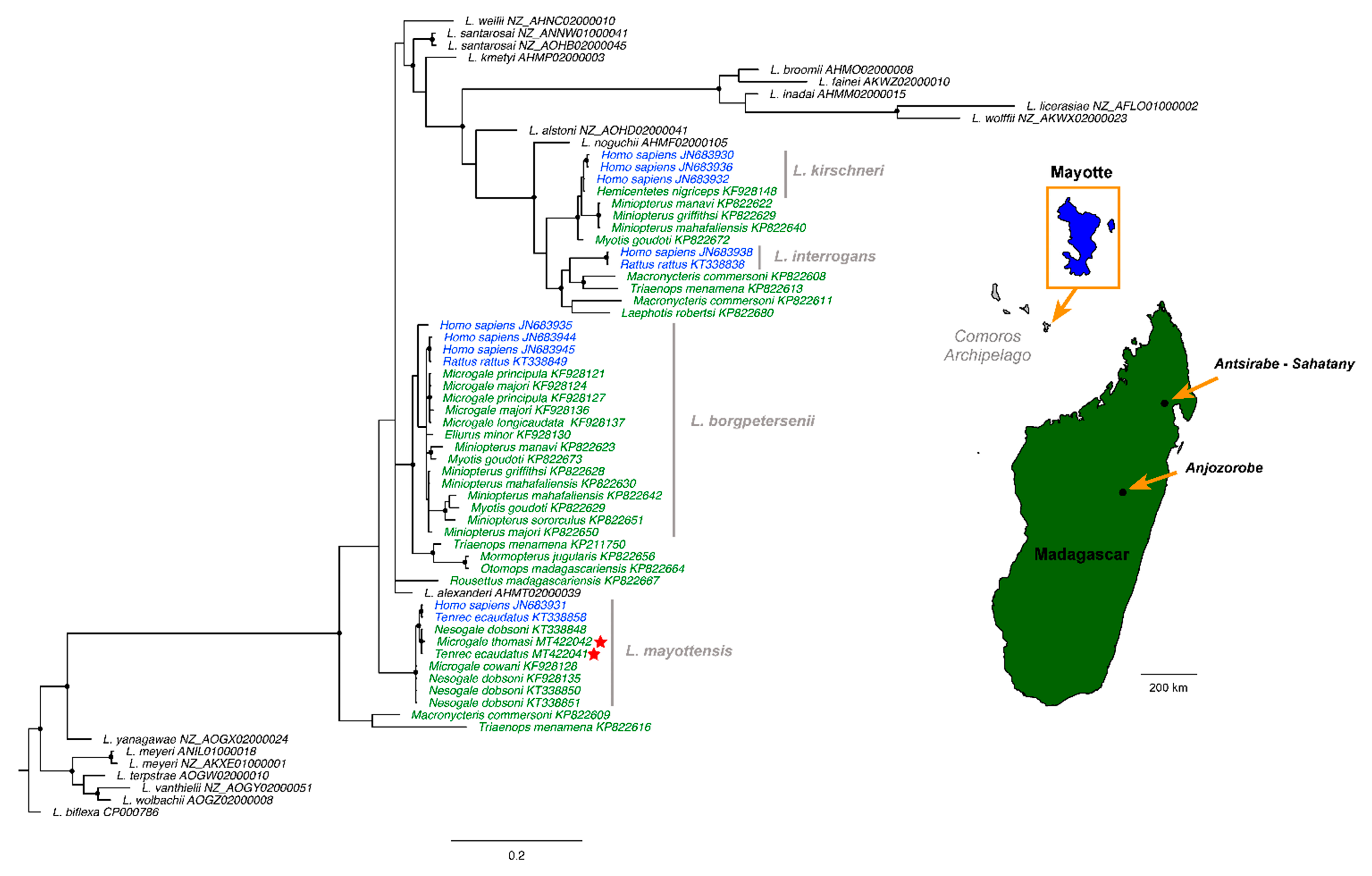
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global morbidity and mortality of leptospirosis: A systematic review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [PubMed]
- Ristow, P.; Bourhy, P.; Kerneis, S.; Schmitt, C.; Prevost, M.-C.; Lilenbaum, W.; Picardeau, M. Biofilm formation by saprophytic and pathogenic leptospires. Microbiology 2008, 154, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Adler, B. History of leptospirosis and Leptospira. Curr. Top. Microbiol. Immunol. 2015, 387, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ko, A.I.; Goarant, C.; Picardeau, M. Leptospira: The dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 2009, 7, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Masuzawa, T.; Sakakibara, K.; Saito, M.; Hidaka, Y.; Villanueva, S.Y.A.M.; Yanagihara, Y.; Yoshida, S. Characterization of Leptospira species isolated from soil collected in Japan. Microbiol. Immunol. 2018, 62, 55–59. [Google Scholar] [CrossRef]
- Thibeaux, R.; Iraola, G.; Ferrés, I.; Bierque, E.; Girault, D.; Soupé-Gilbert, M.-E.; Picardeau, M.; Goarant, C. Deciphering the unexplored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb. Genom. 2018, 4, e000144. [Google Scholar] [CrossRef]
- Thibeaux, R.; Girault, D.; Bierque, E.; Soupé-Gilbert, M.-E.; Rettinger, A.; Douyère, A.; Meyer, M.; Iraola, G.; Picardeau, M.; Goarant, C. Biodiversity of environmental Leptospira: Improving identification and revisiting the diagnosis. Front. Microbiol. 2018, 9, 816. [Google Scholar] [CrossRef]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Khalid, M.K.N.M.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef]
- Bourhy, P.; Collet, L.; Lernout, T.; Zinini, F.; Hartskeerl, R.A.; van der Linden, H.; Thiberge, J.M.; Diancourt, L.; Brisse, S.; Giry, C.; et al. Human Leptospira isolates circulating in Mayotte (Indian Ocean) have unique serological and molecular features. J. Clin. Microbiol. 2012, 50, 307–311. [Google Scholar] [CrossRef]
- Cosson, J.-F.; Picardeau, M.; Mielcarek, M.; Tatard, C.; Chaval, Y.; Suputtamongkol, Y.; Buchy, P.; Jittapalapong, S.; Herbreteau, V.; Morand, S. Epidemiology of Leptospira transmitted by rodents in Southeast Asia. PLoS Negl. Trop. Dis. 2014, 8, e2902. [Google Scholar] [CrossRef]
- Guernier, V.; Lagadec, E.; Cordonin, C.; Minter, G.L.; Gomard, Y.; Pagès, F.; Jaffar-Bandjee, M.-C.; Michault, A.; Tortosa, P.; Dellagi, K. Human leptospirosis on Reunion Island, Indian Ocean: Are rodents the (only) ones to blame? PLoS Negl. Trop. Dis. 2016, 10, e0004733. [Google Scholar] [CrossRef] [PubMed]
- Biscornet, L.; Dellagi, K.; Pagès, F.; Bibi, J.; Comarmond, J.D.; Mélade, J.; Govinden, G.; Tirant, M.; Gomard, Y.; Guernier, V.; et al. Human leptospirosis in Seychelles: A prospective study confirms the heavy burden of the disease but suggests that rats are not the main reservoir. PLoS Negl. Trop. Dis. 2017, 11, e0005831. [Google Scholar] [CrossRef] [PubMed]
- Guernier, V.; Richard, V.; Nhan, T.; Rouault, E.; Tessier, A.; Musso, D. Leptospira diversity in animals and humans in Tahiti, French Polynesia. PLoS Negl. Trop. Dis. 2017, 11, e0005676. [Google Scholar] [CrossRef] [PubMed]
- Gomard, Y.; Dellagi, K.; Goodman, S.M.; Mavingui, P.; Tortosa, P. Tracking animal reservoirs of pathogenic Leptospira: The right test for the right claim. Trop. Med. Infect. Dis. 2021, 6, 205. [Google Scholar] [CrossRef]
- Gomard, Y.; Dietrich, M.; Wieseke, N.; Ramasindrazana, B.; Lagadec, E.; Goodman, S.M.; Dellagi, K.; Tortosa, P. Malagasy bats shelter a considerable genetic diversity of pathogenic Leptospira suggesting notable host-specificity patterns. FEMS Microbiol. Ecol. 2016, 92, fiw037. [Google Scholar] [CrossRef]
- Tortosa, P.; Dellagi, K.; Mavingui, P. Leptospiroses on the French Islands of the Indian Ocean. Bull. Epidemiol. Hebd. 2017, 8–9, 157–161. [Google Scholar]
- Dietrich, M.; Gomard, Y.; Lagadec, E.; Ramasindrazana, B.; Le Minter, G.; Guernier, V.; Benlali, A.; Rocamora, G.; Markotter, W.; Goodman, S.M.; et al. Biogeography of Leptospira in wild animal communities inhabiting the insular ecosystem of the western Indian Ocean islands and neighboring Africa. Emerg. Microbes Infect. 2018, 7, 57. [Google Scholar] [CrossRef]
- Bourhy, P.; Collet, L.; Clément, S.; Huerre, M.; Ave, P.; Giry, C.; Pettinelli, F.; Picardeau, M. Isolation and characterization of new Leptospira genotypes from patients in Mayotte (Indian Ocean). PLoS Negl. Trop. Dis. 2010, 4, e724. [Google Scholar] [CrossRef]
- Bourhy, P.; Collet, L.; Brisse, S.; Picardeau, M. Leptospira mayottensis sp. nov., a pathogenic species of the genus Leptospira isolated from humans. Int. J. Syst. Evol. Microbiol. 2014, 64, 4061–4067. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.; Wilkinson, D.A.; Soarimalala, V.; Goodman, S.M.; Dellagi, K.; Tortosa, P. Diversification of an emerging pathogen in a biodiversity hotspot: Leptospira in endemic small mammals of Madagascar. Mol. Ecol. 2014, 23, 2783–2796. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.; Gomard, Y.; Cordonin, C.; Tortosa, P. Leptospira bacteria in Madagascar. In The New Natural History of Madagascar; Goodman, S.M., Ed.; Princeton University Press: Princeton, NJ, USA, 2022; pp. 268–277. [Google Scholar]
- Everson, K.M.; Soarimalala, V.; Goodman, S.M.; Olson, L.E. Multiple loci and complete taxonomic sampling resolve the phylogeny and biogeographic history of Tenrecs (Mammalia: Tenrecidae) and reveal higher speciation rates in Madagascar’s humid forests. Syst. Biol. 2016, 65, 890–909. [Google Scholar] [CrossRef] [PubMed]
- Goodman, S.M.; Soarimalala, V.; Olson, L.E. Systématique des tenrecs endémiques malgaches (famille des Tenrecidae)/Systematics of endemic Malagasy Tenrecs (family Tenrecidae). In Les Aires Protégées Terrestres de Madagascar: Leur Histoire, Description, et Biote./The Terrestrial Protected Areas of Madagascar: Their History, Description and Biota; Goodman, S.M., Raherilalao, M.J., Wohlauser, S., Eds.; Association Vahatra: Antananarivo, Madagascar, 2018; pp. 363–372. [Google Scholar]
- Lagadec, E.; Gomard, Y.; Minter, G.L.; Cordonin, C.; Cardinale, E.; Ramasindrazana, B.; Dietrich, M.; Goodman, S.M.; Tortosa, P.; Dellagi, K. Identification of Tenrec ecaudatus, a wild mammal introduced to Mayotte Island, as a reservoir of the newly identified human pathogenic Leptospira mayottensis. PLoS Negl. Trop. Dis. 2016, 10, e0004933. [Google Scholar] [CrossRef] [PubMed]
- Annapragada, A.; Brook, C.E.; Luskin, M.S.; Rahariniaina, R.P.; Helin, M.; Razafinarivo, O.; Ambinintsoa Ralaiarison, R.; Randriamady, H.J.; Olson, L.E.; Goodman, S.M.; et al. Evaluation of tenrec population viability and potential sustainable management under hunting pressure in northeastern Madagascar. Anim. Conserv. 2021, 24, 1059–1070. [Google Scholar] [CrossRef]
- Sikes, R.S. The Animal Care and Use Committee of the American Society of Mammalogists. Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J. Mammal. 2016, 97, 663–688. [Google Scholar] [CrossRef]
- Becker, R.A.; Wilks, A.R. R Version by Ray Brownrigg. Mapdata: Extra Map Databases. R Package Version 2.3.0. 2018. Available online: https://cran.r-project.org/web/packages/mapdata/ (accessed on 12 August 2022).
- Smythe, L.D.; Smith, I.L.; Smith, G.A.; Dohnt, M.F.; Symonds, M.L.; Barnett, L.J.; McKay, D.B. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect. Dis. 2002, 2, 13. [Google Scholar] [CrossRef]
- Ahmed, N.; Devi, S.M.; Valverde, M.D.L.A.; Vijayachari, P.; Machang’u, R.S.; Ellis, W.A.; Hartskeerl, R.A. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 2006, 5, 28. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree 1.4.2, a Graphical Viewer of Phylogenetic Trees. 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 12 August 2022).
- Olson, L.E. Tenrecs. Curr. Biol. 2013, 23, R5–R8. [Google Scholar] [CrossRef]
- Emerson, B.C. Evolution on oceanic islands: Molecular phylogenetic approaches to understanding pattern and process. Mol. Ecol. 2002, 11, 951–966. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.W. Washed up in Madagascar. Nature 2010, 463, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Lovegrove, B.G.; Lobban, K.D.; Levesque, D.L. Mammal survival at the Cretaceous–Palaeogene boundary: Metabolic homeostasis in prolonged tropical hibernation in tenrecs. Proc. Biol. Sci. 2014, 281, 20141304. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.A.; Mélade, J.; Dietrich, M.; Ramasindrazana, B.; Soarimalala, V.; Lagadec, E.; Minter, G.L.; Tortosa, P.; Heraud, J.-M.; Lamballerie, X.D.; et al. Highly diverse Morbillivirus-related paramyxoviruses in wild fauna of the southwestern Indian Ocean islands: Evidence of exchange between introduced and endemic small mammals. J. Virol. 2014, 88, 8268–8277. [Google Scholar] [CrossRef]
- Moseley, M.; Rahelinirina, S.; Rajerison, M.; Garin, B.; Piertney, S.; Telfer, S. Mixed Leptospira infections in a diverse reservoir host community, Madagascar, 2013–2015. Emerg. Infect. Dis. 2018, 24, 1138–1140. [Google Scholar] [CrossRef]
- Cordonin, C.; Turpin, M.; Bringart, M.; Bascands, J.-L.; Flores, O.; Dellagi, K.; Mavingui, P.; Roche, M.; Tortosa, P. Pathogenic Leptospira and their animal reservoirs: Testing host specificity through experimental infection. Sci. Rep. 2020, 10, 7239. [Google Scholar] [CrossRef]
- Desvars, A.; Naze, F.; Benneveau, A.; Cardinale, E.; Michault, A. Endemicity of leptospirosis in domestic and wild animal species from Reunion Island (Indian Ocean). Epidemiol. Infect. 2013, 141, 1154–1165. [Google Scholar] [CrossRef]
- Goodman, S.M.; Soarimalala, V. Introduction to mammals. In The New Natural History of Madagascar; Goodman, S.M., Ed.; Princeton University Press: Princeton, NJ, USA, 2022; pp. 1737–1769. [Google Scholar]
- Baas Becking, L.G.M. Geobiologie of Inleiding Tot de Milieukunde; W.P. Van Stockum & Zoon NV: The Hague, The Netherlands, 1934. [Google Scholar]
- De Wit, R.; Bouvier, T. ‘Everything is everywhere, but, the environment Selects’; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 2006, 8, 755–758. [Google Scholar] [CrossRef]
- Fierer, N. Microbial biogeography: Patterns in microbial diversity across space and time. In Accessing Uncultivated Microorganisms: From the Environment to Organisms and Genomes and Back; Zengler, K., Ed.; ASM Press: Washington, WA, USA, 2008; pp. 95–115. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomard, Y.; Goodman, S.M.; Soarimalala, V.; Turpin, M.; Lenclume, G.; Ah-Vane, M.; Golden, C.D.; Tortosa, P. Co-Radiation of Leptospira and Tenrecidae (Afrotheria) on Madagascar. Trop. Med. Infect. Dis. 2022, 7, 193. https://doi.org/10.3390/tropicalmed7080193
Gomard Y, Goodman SM, Soarimalala V, Turpin M, Lenclume G, Ah-Vane M, Golden CD, Tortosa P. Co-Radiation of Leptospira and Tenrecidae (Afrotheria) on Madagascar. Tropical Medicine and Infectious Disease. 2022; 7(8):193. https://doi.org/10.3390/tropicalmed7080193
Chicago/Turabian StyleGomard, Yann, Steven M. Goodman, Voahangy Soarimalala, Magali Turpin, Guenaëlle Lenclume, Marion Ah-Vane, Christopher D. Golden, and Pablo Tortosa. 2022. "Co-Radiation of Leptospira and Tenrecidae (Afrotheria) on Madagascar" Tropical Medicine and Infectious Disease 7, no. 8: 193. https://doi.org/10.3390/tropicalmed7080193
APA StyleGomard, Y., Goodman, S. M., Soarimalala, V., Turpin, M., Lenclume, G., Ah-Vane, M., Golden, C. D., & Tortosa, P. (2022). Co-Radiation of Leptospira and Tenrecidae (Afrotheria) on Madagascar. Tropical Medicine and Infectious Disease, 7(8), 193. https://doi.org/10.3390/tropicalmed7080193





