Infection of Rodents by Orientia tsutsugamushi, the Agent of Scrub Typhus in Relation to Land Use in Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Study Sites and Rodent Trappings
2.3. Environmental Indices and Land Use
2.4. Rodent Screening for O. tsutsugamushi
2.5. Statistical Analyses
3. Results
3.1. Micro-Mammal Identification and O. tsutsugamushi Screening
3.2. Individual Surrounding Habitat Characteristics and O. tsutsugamushi Infection
4. Discussion
4.1. Limitations of Our Study
4.2. Implications for Public Health
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kelly, D.J.; Fuerst, P.A.; Ching, W.M.; Richards, A.L. Scrub typhus: The geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 2009, 15, S203–S230. [Google Scholar] [CrossRef] [PubMed]
- Cosson, J.-F.; Galan, M.; Bard, E.; Razzauti, M.; Bernard, M.; Morand, S.; Brouat, C.; Dalecky, A.; Ba, K.; Charbonnel, N.; et al. Detection of Orientia sp. DNA in rodents from Asia, West Africa and Europe. Parasites Vect. 2015, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Watt, G.; Parola, P. Scrub typhus and tropical rickettsioses. Curr. Opinion Infect. Dis. 2003, 16, 429–436. [Google Scholar] [CrossRef]
- Walker, M.D. Scrub typhus—Scientific neglect, ever-widening impact. N. Engl. J. Med. 2016, 375, 913–915. [Google Scholar] [CrossRef] [PubMed]
- Chaisiri, K.; McGarry, J.W.; Morand, S.; Makepeace, B.L. Symbiosis in an overlooked microcosm: A systematic review of the bacterial flora of mites. Parasitology 2015, 142, 1152–1156. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.W.; Rapmund, G.; Gadigan, F.G. Sex ratios in Rickettsia tsutsugamushi-infected and noninfected colonies of Leptotrombidium. (Acari: Trombiculidae). J. Med. Entomol. 1977, 14, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Frances, S.P.; Watcharapichat, P.; Phulsuksombati, D. Vertical transmission of Orientia tsutsugamushi in two lines of naturally infected Leptotrombidium deliense (Acari: Trombiculidae). J. Med. Entomol. 2001, 38, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Phasomkusolsil, S.; Tanskul, P.; Ratanatham, S.; Watcharapichat, P.; Phulsuksombati, D.; Frances, S.P.; Lerdthusnee, K.; Linthicum, K.J. Influence of Orientia tsutsugamushi infection on the developmental biology of Leptotrombidium imphalum and Leptotrombidium chiangraiensis (Acari: Trombiculidae). J. Med. Entomol. 2012, 49, 1270–1275. [Google Scholar] [CrossRef]
- Traub, R.; Wisseman, C.L.J. The ecology of chigger-borne rickettsiosis (scrub typhus). J. Med. Entomol. 1974, 11, 237–303. [Google Scholar] [CrossRef] [PubMed]
- Stekolnikov, A.A. Leptotrombidium (Acari: Trombiculidae) of the world. Zootaxa 2013, 3728, zootaxa.3728.1.1. [Google Scholar] [CrossRef]
- Santibanez, P.; Palomar, A.M.; Portillo, A.; Santibanez, S.; Oteo, J.A. The role of chiggers as human pathogens. Available online: https://www.intechopen.com/books/an-overview-of-tropical-diseases/the-role-of-chiggers-as-human-pathogens (accessed on 5 October 2017).
- Coleman, R.E.; Monkanna, T.; Linthicum, K.J.; Strickman, D.A.; Frances, S.P.; Tanskul, P.; Kollars, T.M., Jr.; Inlao, I.; Watcharapichat, P.; Khlaimanee, N.; et al. Occurrence of Orientia tsutsugamushi in small mammals from Thailand. Am. J. Trop. Med. Hyg. 2003, 69, 519–524. [Google Scholar] [PubMed]
- Paris, D.H.; Shelite, T.R.; Day, N.P.; Walker, D.H. Unresolved problems related to scrub typhus: A seriously neglected life-threatening disease. Am. J. Trop. Med. Hyg. 2013, 89, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Shatrov, A.B.; Kudryashova, N.I. Taxonomic ranking of major trombiculid subtaxa with remarks on the evolution of host-parasite relationships (Acariformes: Parasitengona: Trombiculidae). Ann. Zool. 2008, 58, 279–287. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, Z.; Yang, H.L.; Zhang, A.H.; Xu, X.Q.; Meng, X.P.; Zhang, H.Y.; Wang, X.J.; Li, Z.; Ding, S.J.; et al. Molecular epidemiology of Orientia tsutsugamushi in chiggers and ticks from domestic rodents in Shandong, northern China. Parasites Vect. 2013, 6, 312. [Google Scholar] [CrossRef] [PubMed]
- Chaisiri, K.; Stekolnikov, A.A.; Makepeace, B.L.; Morand, S. A Revised checklist of chigger mites (Acari: Trombiculidae) from Thailand, with the description of three new species. J. Med. Entomol. 2016, 53, 321–342. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.Y.; Guo, X.G.; Ren, T.G.; Song, W.Y.; Dong, W.G.; Fan, R. Species diversity of ectoparasitic chigger mites (Acari: Prostigmata) on small mammals in Yunnan Province, China. Parasitol. Res. 2016, 15, 3605–3618. [Google Scholar] [CrossRef] [PubMed]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009, 35, 221–270. [Google Scholar] [CrossRef] [PubMed]
- Khuntirat, B.; Lerdthusnee, K.; Leepitakrat, W.; Kengleucha, A.; Wongkalasin, K.; Monkanna, T.; Mungviriya, S.; Jones, J.W.; Coleman, R.E. Characterization of Orientia tsutsugamushi isolated from wild-caught rodents and chiggers in northern Thailand. Ann. N. Y. Acad. Sci. 2003, 990, 205–212. [Google Scholar] [CrossRef]
- Lerdthusnee, K.; Nigro, J.; Monkanna, T.; Leepitakrat, W.; Leepitakrat, S.; Insuan, S.; Charoensongsermkit, W.; Khlaimanee, N.; Akkagraisee, W.; Chayapum, K.; et al. Surveys of rodent-borne disease in Thailand with a focus on scrub typhus assessment. Integrat. Zool. 2008, 3, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Wangroongsarb, P.; Saengsongkong, W.; Petkanjanapong, W.; Mimgratok, M.; Panjai, D.; Wootta, W.; Hagiwara, T. An application of duplex PCR for detection of Leptospira spp. and Orientia tsutsugamushi from wild rodents. Jpn. J. Inf. Dis. 2008, 61, 407–409. [Google Scholar]
- Rodkvamtook, W.; Gaywee, J.; Kanjanavanit, S.; Ruangareerate, T.; Richards, A.L.; Sangjun, N.; Jeamwattanalert, P.; Sirisopana, N. Scrub typhus outbreak, northern Thailand, 2006–2007. Emerg. Inf. Dis. 2013, 19, 774–777. [Google Scholar] [CrossRef] [PubMed]
- Rodkvamtook, W.; Ruang-Areerate, T.; Gaywee, J.; Richards, A.L.; Jeamwattanalert, P.; Bodhidatta, D.; Sangjun, N.; Prasartvit, A.; Jatisatienr, A.; Jatisatienr, C. Isolation and Characterization of Orientia tsutsugamushi from rodents captured following a scrub typhus outbreak at a military training base, Bothong District, Chonburi Province, Central Thailand. Am. J. Trop. Med. Hyg. 2011, 84, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Herbreteau, V.; Bordes, F.; Jittapalapong, S.; Supputamongkol, Y.; Morand, S. Rodent-borne diseases in Thailand: Targeting rodent carriers and risky habitats. Infect. Ecol. Epidemiol. 2012, 2, 18637. [Google Scholar] [CrossRef] [PubMed]
- Chareonviriyaphap, T.; Leepitakrat, W.; Lerdthusnee, K.; Chao, C.C.; Ching, W.M. Dual exposure of Rickettsia typhi and Orientia tsutsugamushi in the field-collected Rattus rodents from Thailand. J. Vect. Ecol. 2014, 39, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Knight, R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009, 19, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Muul, I.; Lim, B.L.; Walker, J.S. Scrub typhus infection in rats in four habitats in Peninsular Malaysia. Trans. R Soc. Trop. Med. Hyg. 1977, 71, 493–497. [Google Scholar] [CrossRef]
- Wardrop, N.A.; Kuo, C.-C.; Wang, H.-C.; Clements, A.C.A.; Lee, P.-F.; Atkinson, P.M. Bayesian spatial modelling and the significance of agricultural land use to scrub typhus infection in Taiwan. Geospatial Health 2013, 8, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Morand, S.; Bordes, F.; Blasdell, K.; Pilosof, S.; Cornu, J.-F.; Chaisiri, K.; Chaval, Y.; Cosson, J.-F.; Claude, J.; Feyfant, T.; et al. Assessing the distribution of disease-bearing rodents in human-modified tropical landscapes. J. Appl. Ecol. 2015, 52, 784–794. [Google Scholar] [CrossRef]
- Sun, Y.; Wei, Y.-H.; Yang, Y.; Ma, Y.; de Vlas, S.J.; Yao, H.W.; Huang, Y.; Ma, M.J.; Liu, K.; Li, X.N.; et al. Rapid increase of scrub typhus incidence in Guangzhou, southern China, 2006–2014. BMC Inf. Dis. 2017, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.C.; Huang, J.L.; Shu, P.Y.; Lee, P.L.; Kelt, D.A.; Wang, H.C. Cascading effect of economic globalization on human risks of scrub typhus and tick-borne rickettsial diseases. Ecol. Appl. 2012, 22, 1803–1816. [Google Scholar] [CrossRef] [PubMed]
- Bordes, F.; Morand, S.; Pilosof, S.; Claude, J.; Cosson, J.-F.; Chaval, Y.; Ribas, A.; Chaisiri, K.; Blasdell, K.; Tran, A. Habitat fragmentation alters the properties of a host-parasite network: Rodents and their helminths in South-East Asia. J. Anim. Ecol. 2015, 84, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- Bordes, F.; Caron, A.; Blasdell, K.; de Garine Wichatitsky, M.; Morand, S. Forecasting potential emergence of zoonotic diseases in South-East Asia: Network analysis identifies key rodent hosts. J. Appl. Ecol. 2016, 54. [Google Scholar] [CrossRef]
- Herbreteau, V.; Rerkamnuaychoke, W.; Jittapalapong, S.; Chaval, Y.; Cosson, J.-F.; Morand, S. Field and laboratory protocols for rodent studies. Kasetsart University. Available online: www.ceropath.org/FichiersComplementaires/Herbreteau_Rodents_protocols_2011.pdf (accessed on 6 October 2017).
- Bureau of Epidemiology, Department of Disease Control, Ministry of Public Health, Thailand. Annual epidemiologic surveillance report 2007. Scrub typhus. Available online: http://www.boe.moph.go.th/Annual/ANNUAL2550/Part1/Annual_MenuPart1.html (accessed on 4 October 2017).
- Galan, M.; Pagès, M.; Cosson, J.F. Next-generation sequencing for rodent barcoding: Species identification from fresh, degraded and environmental samples. PLoS ONE 2012, 7, e4837. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Wang, Q.; O’Sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.P.; O’Toole, P.W. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Env. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Silva. Available online: http://www.arb-silva.de/projects/ssu-ref-nr/ (accessed on 6 October 2017).
- Galan, M.; Razzauti, M.; Bard, E.; Bernard, M.; Brouat, C.; Charbonnel, N.; Dehne-Garcia, A.; Loiseau, A.; Tatard, C.; Tamisier, L.; et al. 16S rRNA amplicon sequencing for epidemiological surveys of bacteria in wildlife. MSystems 2016, 1, e00032-16. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Software 2015, 67, 1–48. [Google Scholar] [CrossRef]
- R Development Core Team. The R project for statistical computing, R version 3.4.1, 2017. Available online: https://www.r-project.org (accessed on 4 October 2017).
- Calcagno, V.; de Mazancourt, C. Glmulti: An R package for easy automated model selection with (generalized) linear models. J. Stat. Software 2010, 34, 12. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by random Forest. R News 2002, 2, 18–22. [Google Scholar]
- Bordes, F.; Blasdell, K.; Morand, S. Transmission ecology of rodent-borne diseases: New frontiers. Integrat. Zool. 2015, 10, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Kosoy, M.; Khlyap, L.; Cosson, J.-F.; Morand, S. Aboriginal and invasive rats of genus Rattus as hosts of infectious agents. Vector-Borne Zoon Dis. 2015, 15, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Wilcove, D.S.; Giam, X.; Edwards, D.P.; Fisher, B.; Koh, L.P. Navjot’s nightmare revisited: Logging, agriculture, and biodiversity in Southeast Asia. Trends Ecol. Evol. 2013, 28, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Cornu, J.-F.; Lajaunie, C.; Laborde, H.; Morand, S. Landscape changes and policies for biodiversity and environment conservation in Southeast Asia. In Biodiversity Conservation in Southeast Asia: Challenges in a Changing Environment; Morand, S., Lajaunie, C., Satrawaha, R., Eds.; Routledge: Abingdon, UK, 2017; pp. 49–66. [Google Scholar]
- Razzauti, M.; Galan, M.; Bernard, M.; Maman, S.; Klopp, C.; Charbonnel, N.; Vayssier-Taussat, M.; Eloit, M.; Cosson, J.F. A comparison between transcriptome sequencing and 16S metagenomics for detection of bacterial pathogens in wildlife. PLoS Negl. Trop. Dis. 2015, 9, e0003929. [Google Scholar] [CrossRef] [PubMed]
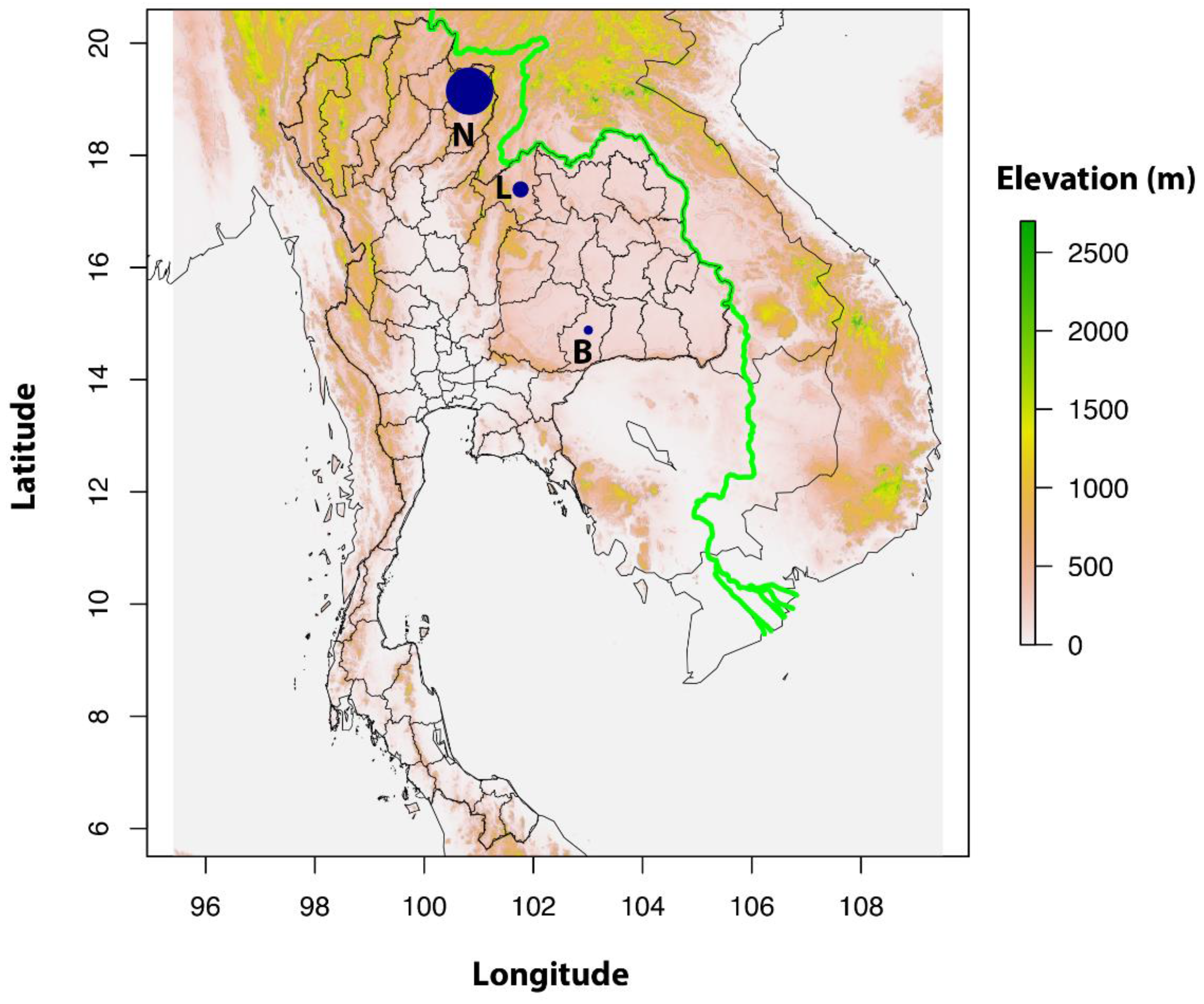
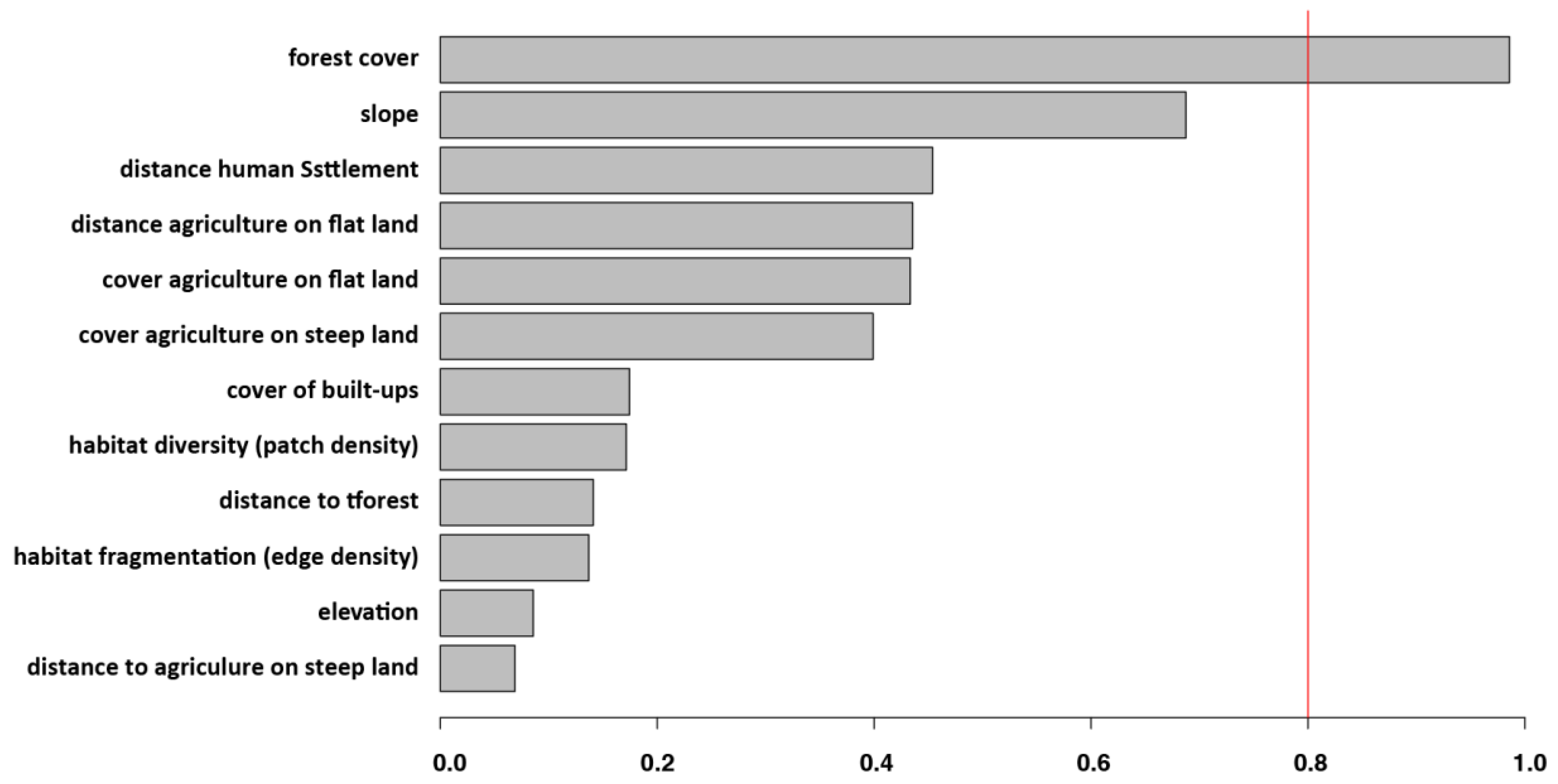
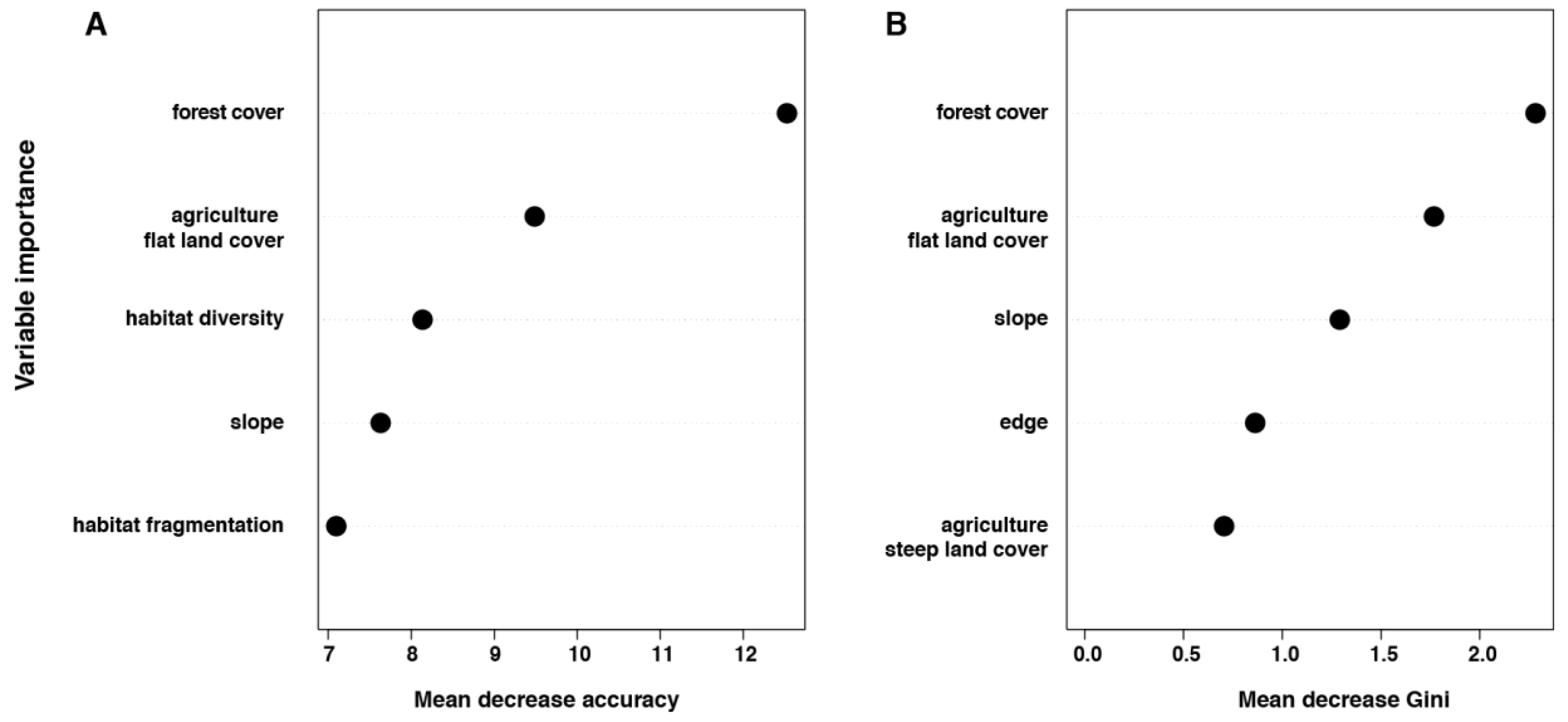
| Locality | Micro-Mammal Species | Number Tested | O. tsutsugamushi Positive |
|---|---|---|---|
| Buriram | Bandicota indica | 2 | 0 |
| Bandicota savilei | 10 | 2 | |
| Menetes berdmorei | 1 | 0 | |
| Mus caroli | 8 | 0 | |
| Mus cervicolor | 11 | 0 | |
| Mus cookii | 1 | 0 | |
| Rattus argentiventer | 1 | 0 | |
| Rattus exulans | 32 | 0 | |
| Rattus sakaretensis | 2 | 0 | |
| Rattus tanezumi | 3 | 1 | |
| Rattus sp. clade 3 | 26 | 2 | |
| Loei | Bandicota indica | 15 | 0 |
| Bandicota savilei | 12 | 0 | |
| Berylmys berdmorei | 9 | 0 | |
| Berylmys bowersi | 14 | 1 | |
| Cannomys badius | 1 | 0 | |
| Chiropodomys gliroides | 2 | 0 | |
| Crocidura attenuate | 1 | 0 | |
| Hapalomys delacouri | 1 | 0 | |
| Leopoldamys edwardsi | 5 | 1 | |
| Leopoldamys sabanus | 5 | 0 | |
| Maxomys surifer | 15 | 0 | |
| Mus caroli | 5 | 0 | |
| Mus cervicolor | 5 | 0 | |
| Mus cookii | 6 | 0 | |
| Mus fragilicauda | 1 | 0 | |
| Niviventer fulvescens | 16 | 0 | |
| Rattus exulans | 26 | 1 | |
| Rattus sakaretensis | 30 | 0 | |
| Rattus nitidus | 1 | 0 | |
| Rattus tanezumi | 16 | 0 | |
| R. tanezumi clade 3 | 2 | 0 | |
| Nan | Bandicota indica | 30 | 0 |
| Berylmys berdmorei | 8 | 0 | |
| Berylmys bowersi | 1 | 0 | |
| Mus caroli | 1 | 0 | |
| Mus cookiie | 4 | 0 | |
| Rattus exulans | 24 | 0 | |
| Rattus tanezumi | 25 | 1 |
| Models (Best Top Three) | K | AICc | wr |
|---|---|---|---|
| slope + cover of forest + distance to agriculture on flat land | 4 | 84.70 | 0.033 |
| slope + cover of forest | 3 | 85.02 | 0.028 |
| Slope+ cover of agriculture on steep land + cover of agriculture on flat land + cover of forest + distance to agriculture on flat land + distance to human settlement | 6 | 85.22 | 0.025 |
| Explanatory Surrounding Habitat Characteristics | Estimate (SD), p | Log Likelihood, Deviance (DF) |
|---|---|---|
| Forest cover | 3.87 (1.25), 0.002 | |
| Distance to agriculture on flat land | 0.003 (0.001), 0.096 | |
| Slope | −0.22 (0.14), 0.10 | −160.6, 321.1 (1360) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaisiri, K.; Cosson, J.-F.; Morand, S. Infection of Rodents by Orientia tsutsugamushi, the Agent of Scrub Typhus in Relation to Land Use in Thailand. Trop. Med. Infect. Dis. 2017, 2, 53. https://doi.org/10.3390/tropicalmed2040053
Chaisiri K, Cosson J-F, Morand S. Infection of Rodents by Orientia tsutsugamushi, the Agent of Scrub Typhus in Relation to Land Use in Thailand. Tropical Medicine and Infectious Disease. 2017; 2(4):53. https://doi.org/10.3390/tropicalmed2040053
Chicago/Turabian StyleChaisiri, Kittipong, Jean-François Cosson, and Serge Morand. 2017. "Infection of Rodents by Orientia tsutsugamushi, the Agent of Scrub Typhus in Relation to Land Use in Thailand" Tropical Medicine and Infectious Disease 2, no. 4: 53. https://doi.org/10.3390/tropicalmed2040053
APA StyleChaisiri, K., Cosson, J.-F., & Morand, S. (2017). Infection of Rodents by Orientia tsutsugamushi, the Agent of Scrub Typhus in Relation to Land Use in Thailand. Tropical Medicine and Infectious Disease, 2(4), 53. https://doi.org/10.3390/tropicalmed2040053






