Genotype Analysis on Orientia tsutsugamushi Causing Scrub Typhus in Malaysia: A Re-Emerging Disease
Abstract
1. Introduction
2. Materials and Methods
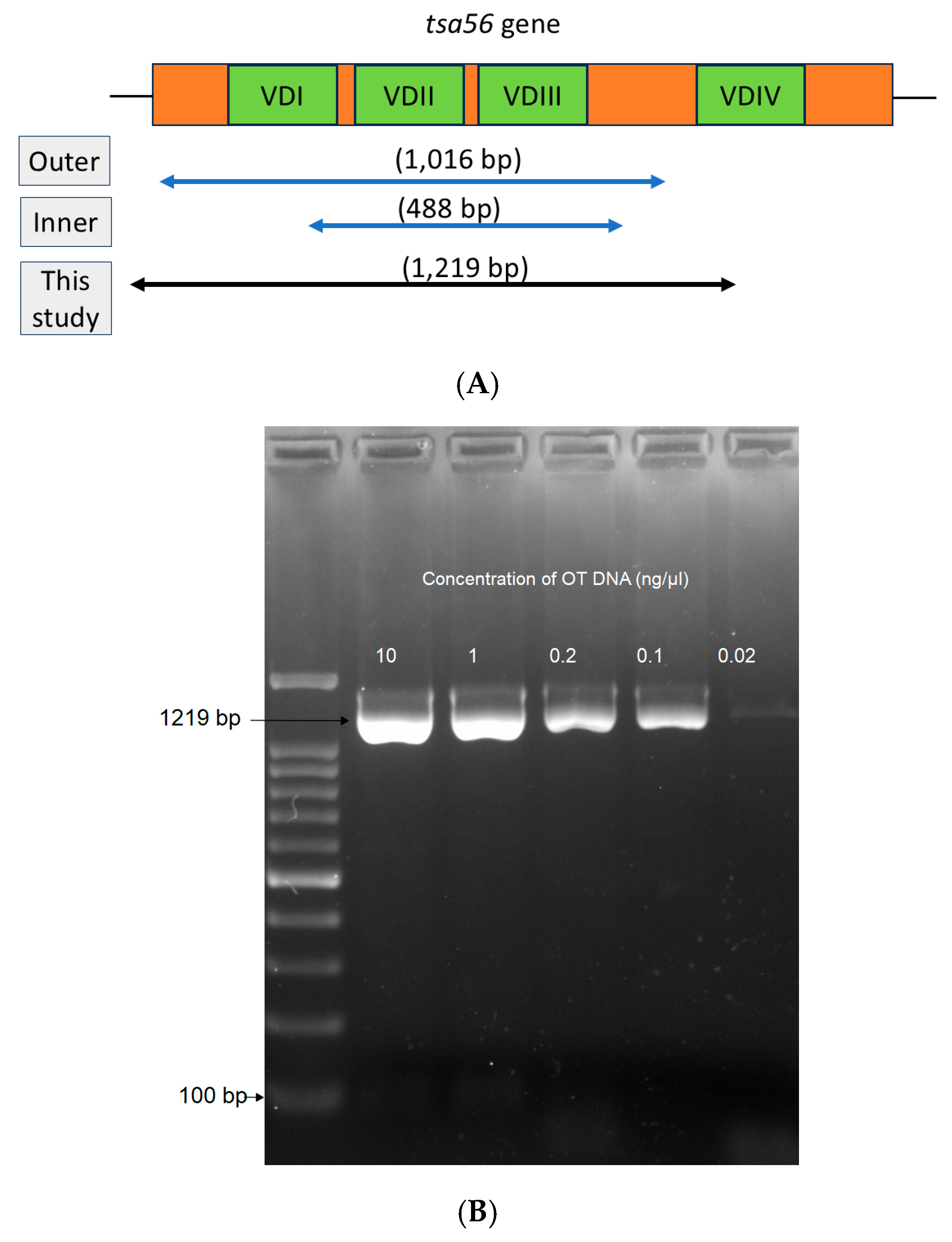
2.1. DNA Extraction and PCR
2.2. Single-Run PCR
2.3. OT Genotyping
2.4. Ethics Statement
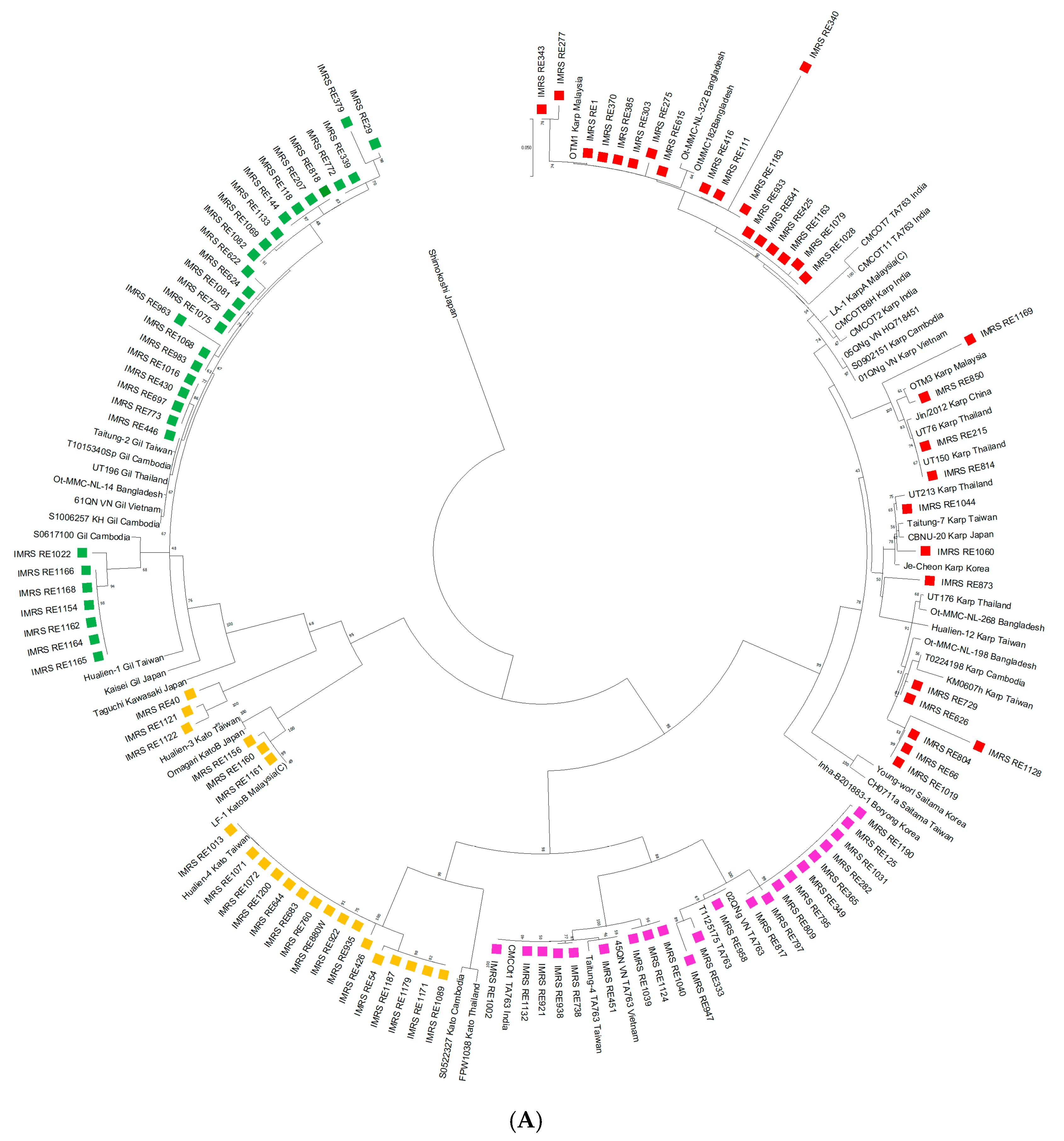
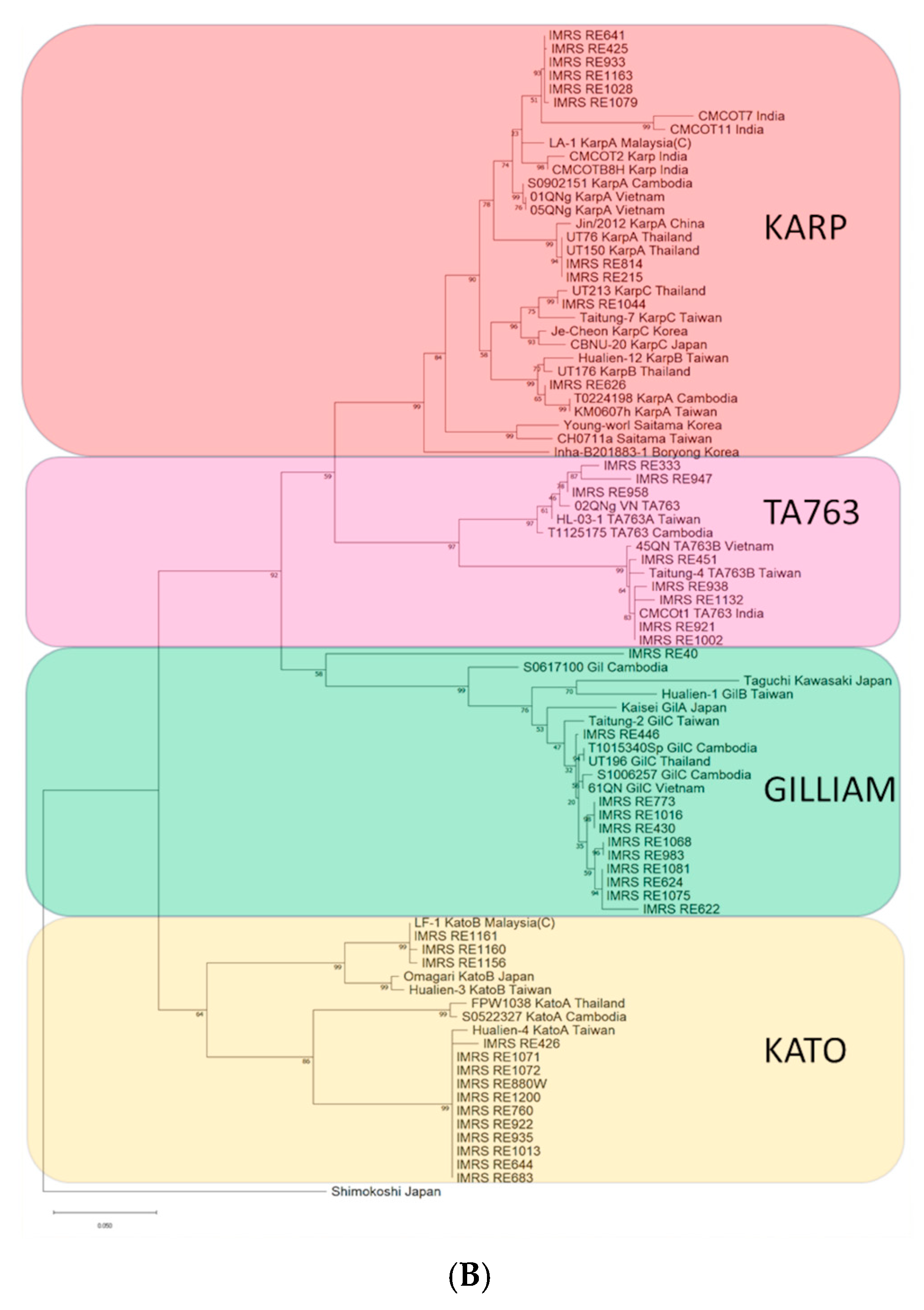
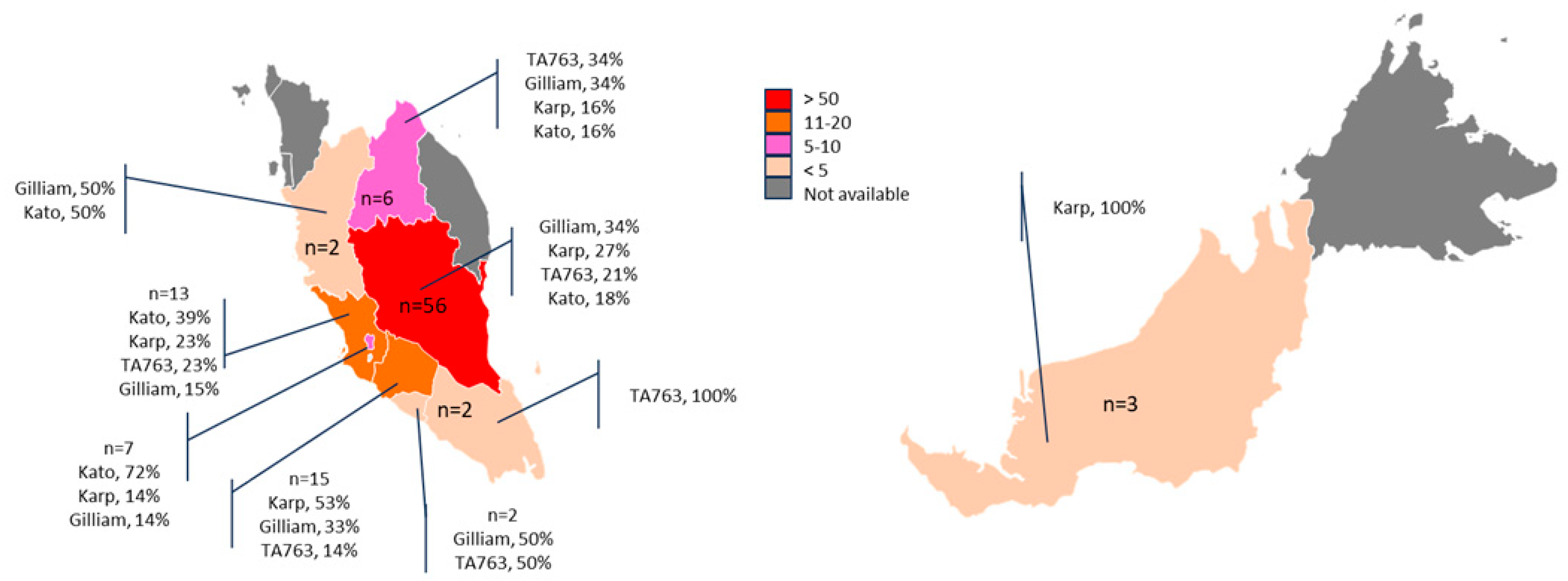
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, G.; Walker, D.H.; Jupiter, D.; Melby, P.C.; Arcari, C.M. A review of the global epidemiology of scrub typhus. PLoS Negl. Trop. Dis. 2017, 11, e0006062. [Google Scholar] [CrossRef]
- Watt, G.; Parola, P. Scrub typhus and tropical rickettsioses. Curr. Opin. Infect. Dis. 2003, 16, 429–436. [Google Scholar] [CrossRef]
- Huxsoll, D.L. Scrub typhus: A problem in Malaysia. Malays. J. Path. 1978, 1, 1–6. [Google Scholar]
- Grigg, M.J.; William, T.; Clemens, E.G.; Patel, K.; Chandna, A.; Wilkes, C.S.; Barber, B.E.; Anstey, N.M.; Dumler, J.S.; Yeo, T.W.; et al. Rickettsioses as Major Etiologies of Unrecognized Acute Febrile Illness, Sabah, East Malaysia. Emerg. Infect. Dis. 2020, 26, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, W.; Field, J.W. The Tsutsugamushi Disease in the Federated Malay States; Bulletin from the Institute for Medical Research, Federated Malay States No.1; Institute for Medical Research: Kuala Lumpur, Malaysia, 1927. [Google Scholar]
- Tay, S.T.; Ho, T.M.; Rohani, M.Y.; Devi, S. Antibodies to Orientia tsutsugamushi, Rickettsia typhi and spotted fever group rickettsiae among febrile patients in rural areas of Malaysia. Trans. R. Soc. Trop. Med. Hyg. 2000, 94, 280–284. [Google Scholar] [CrossRef]
- Yuhana, M.Y.; Hanboonkunupakarn, B.; Tanganuchitcharnchai, A.; Sujariyakul, P.; Sonthayanon, P.; Chotivanich, K.; Pukrittayakamee, S.; Blacksell, S.D.; Paris, D.H. Rickettsial Infections Are Neglected Causes of Acute Febrile Illness in Teluk Intan, Peninsular Malaysia. Trop. Med. Infect. Dis. 2022, 7, 77, Erratum in Trop. Med. Infect. Dis. 2022, 7, 134. https://doi.org/10.3390/tropicalmed7070134. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.A.; Lee, K.S.; Thangavelu, S.; Ng, T.K. Scrub typhus, the forgotten acute febrile illness: A case series from Negeri Sembilan, Malaysia. MJM Case Rep. J. 2024, 3. [Google Scholar] [CrossRef]
- Kim, G.; Ha, N.Y.; Min, C.K.; Kim, H.I.; Yen, N.T.H.; Lee, K.H.; Oh, I.; Kang, J.-S.; Choi, M.-S.; Kim, I.-S.; et al. Diversification of Orientia tsutsugamushi genotypes by intragenic recombination and their potential expansion in endemic areas. PLoS Negl. Trop. Dis. 2017, 11, e0005408. [Google Scholar] [CrossRef]
- Kumaraswamy, J.; Govindasamy, P.; Nagarajan, L.S.; Gunasekaran, K.; Abhilash, K.P.P.; Prakash, J.A.J. Genotyping of Orientia tsutsugamushi circulating in and around Vellore (South India) using TSA 56 gene. Indian J. Med. Microbiol. 2024, 47, 100483. [Google Scholar] [CrossRef]
- Enatsu, T.; Urakami, H.; Tamura, A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol. Lett. 1999, 180, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Bora, T.; Khan, S.A.; Jampa, L.; Laskar, B. Genetic diversity of Orientia tsutsugamushi strains circulating in Northeast India. Trans. R. Soc. Trop. Med. Hyg. 2018, 112, 22–30. [Google Scholar] [CrossRef]
- Rungrojn, A.; Batty, E.M.; Perrone, C.; Abdad, M.Y.; Wangrangsimakul, T.; Brummaier, T.; McGready, R.; Day, N.P.J.; Blacksell, S.D. Molecular diagnosis and genotyping of Orientia tsutsugamushi in Maesot and Chiangrai, Thailand. Front Trop Dis. 2023, 4, 1146138. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jiang, J.; Chan, T.C.; Temenak, J.J.; Dasch, G.A.; Ching, W.M.; Richards, A.L. Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. Am. J. Trop. Med. Hyg. 2004, 70, 351–356. [Google Scholar] [CrossRef]
- Ramli, S.R.; Arifin, N.; Ismail, M.F.; Hii, S.Y.F.; Sulaiman, N.S.; Lah, E.F.C.; Nik Abdul Aziz, N.A.H. Severe Scrub Typhus with Acute Kidney Injury: Urine PCR Evidence from an East Coast Malaysian Cluster. Trop. Med. Infect. Dis. 2025, 10, 208. [Google Scholar] [CrossRef]
- Furuya, Y.; Yoshida, Y.; Katayama, T.; Yamamoto, S.; Kawamura, A., Jr. Serotype-specific amplification of Rickettsia tsutsugamushi DNA by nested polymerase chain reaction. J. Clin. Microbiol. 1993, 31, 1637–1640. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evolut. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Blacksell, S.D.; Luksameetanasan, R.; Kalambaheti, T.; Aukkanit, N.; Paris, D.H.; McGready, R.; Nosten, F.; Peacock, S.J.; Day, N.P. Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol. Med. Microbiol. 2008, 52, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.J.; Fuerst, P.A.; Ching, W.M.; Richards, A.L. Scrub typhus: The geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 2009, 48 (Suppl. S3), S203–S230. [Google Scholar] [CrossRef] [PubMed]
- Sonthayanon, P.; Chierakul, W.; Wuthiekanun, V.; Phimda, K.; Pukrittayakamee, S.; Day, N.P.; Peacock, S.J. Association of high Orientia tsutsugamushi DNA loads with disease of greater severity in adults with scrub typhus. J. Clin. Microbiol. 2009, 47, 430–434. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Al Amin, M.M.; Paul, S.K.; Aung, M.S.; Paul, A.; Aziz, M.A.; Khan, N.A.; Haque, A.; Ahamed, F.; Melan, A.; Sarker, S.; et al. Molecular characterization of Orientia tsutsugamushi causing scrub typhus among febrile patients in north-central Bangladesh. New Microbes New Infect. 2019, 32, 100595. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nila, S.S.; Kobayashi, N.; Ohashi, N.; Paul, S.K.; Nasreen, S.A.; Roy, S.; Ahmed, S.; Khanam, J.; Bin Sayeed, M.A.A.; Paul, S.; et al. Epidemiological features of scrub typhus and molecular characteristics of Orientia tsutsugamushi in north-central Bangladesh. IJID Reg. 2025, 14, 100571. [Google Scholar] [CrossRef]
- Mohamed Zan, H.A.; Sasheela, P.; Omar, S.F.S.; Savithiri, P.D.; Lim, Y.A.; Sun, T.T. Genetic variants of Orientia tsutsugamushi identified from scrub typhus cases in Malaysia. Trop. Biomed. 2016, 33, 203–208. [Google Scholar] [PubMed]
- Tay, S.T.; Rohani, Y.M.; Ho, T.M.; Shamala, D. Sequence analysis of the hypervariable regions of the 56 kDa immunodominant protein genes of Orientia tsutsugamushi strains in Malaysia. Microbiol. Immunol. 2005, 49, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Ernieenor, F.C.L.; NorJaiza, M.J.; Fadillah, A.; Canedy, J.; Mariana, A. Screening and genotyping of Orientia tsutsugamushi from field-collected on-host chiggers (Acari: Prostigmata) recovered from a positive scrub typhus locality in Kelantan, Malaysia. Exp. Appl. Acarol. 2021, 84, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Ha, N.Y.; Ryu, B.; Bang, J.H.; Song, H.; Kim, Y.; Kim, G.; Oh, M.-d.; Cho, N.-H.; Lee, J.-k.; et al. Urbanization of scrub typhus disease in South Korea. PLoS Negl. Trop. Dis. 2015, 9, e0003814. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Blacksell, S.D.; Bryant, N.J.; Paris, D.H.; Doust, J.A.; Sakoda, Y.; Day, N.P. Scrub typhus serologic testing with the indirect immunofluorescence method as a diagnostic gold standard: A lack of consensus leads to a lot of confusion. Clin. Infect. Dis. 2007, 44, 391–401. [Google Scholar] [CrossRef]
- Wang, G.; Fu, R.; Zhang, L.; Xue, L.; Al-Mahdi, A.Y.; Xie, X.; Qin, A.; Tang, C.; Du, J.; Huang, Y.; et al. Genomic bacterial load associated with bacterial genotypes and clinical characteristics in patients with scrub typhus in Hainan Island, Southern China. PLoS Negl. Trop. Dis. 2023, 17, e0011243. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, D.M.; Yun, N.R.; Neupane, G.P.; Shin, S.H.; Ryu, S.Y.; Yoon, H.J.; Wie, S.H.; Kim, W.J.; Lee, C.Y.; Choi, J.S.; et al. Differences in clinical features according to Boryoung and Karp genotypes of Orientia tsutsugamushi. PLoS ONE 2011, 6, e22731. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Demography | n = 106 (%) |
| i. Age group | |
| 0–10 | 1 (0.9) |
| 11–20 | 7 (6.6) |
| 21–30 | 17 (16.1) |
| 31–40 | 24 (22.7) |
| 41–50 | 19 (17.9) |
| 51–60 | 23 (21.7) |
| >60 | 15 (14.1) |
| ii. Gender | |
| Male | 77 (72.6) |
| Female | 29 (27.4) |
| iii. Ethnicity | |
| Malay | 65 (61.3) |
| Chinese | 9 (8.4) |
| Aborigine * | 6 (5.7) |
| Indian | 6 (5.7) |
| Foreigner | 20 (18.9) |
| iv. Residing state | |
| Pahang | 56 (52.8) |
| Negeri Sembilan | 15 (14.1) |
| Selangor | 13 (12.3) |
| Kuala Lumpur | 7 (6.6) |
| Kelantan | 6 (5.7) |
| Sarawak | 3 (2.8) |
| Johor | 2 (1.9) |
| Melaka | 2 (1.9) |
| Perak | 2 (1.9) |
| Clinical features | n = 106 (%) |
| Fever Vomiting | 99 (93) |
| Headache | 42(40) |
| Eschar | 42 (40) |
| Nause | 32 (30) |
| Diarrhea | 17 (16) |
| Rash | 12 (11) |
| Myalgia | 11 (10) |
| Lethargy | 11 (10) |
| Lymphadenopathy | 11 (10) |
| Cough | 8 (8) |
| Poor appetite | 7 (7) |
| Dizziness | 6 (6) |
| Abdominal pain | 5 (5) |
| Sample type | n = 106 (%) |
| Blood | 90 (85) |
| Eschar | 14 (13.2) |
| CSF | 1 (0.9) |
| Urine | 1 (0.9) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hii, S.Y.F.; Mohd Zaidi, M.N.; Kassim, W.N.; Hashim, R.; Ramli, S.R. Genotype Analysis on Orientia tsutsugamushi Causing Scrub Typhus in Malaysia: A Re-Emerging Disease. Trop. Med. Infect. Dis. 2025, 10, 252. https://doi.org/10.3390/tropicalmed10090252
Hii SYF, Mohd Zaidi MN, Kassim WN, Hashim R, Ramli SR. Genotype Analysis on Orientia tsutsugamushi Causing Scrub Typhus in Malaysia: A Re-Emerging Disease. Tropical Medicine and Infectious Disease. 2025; 10(9):252. https://doi.org/10.3390/tropicalmed10090252
Chicago/Turabian StyleHii, Shirley Yi Fen, Maswani Nabilah Mohd Zaidi, Wan Norazanin Kassim, Rohaidah Hashim, and Siti Roszilawati Ramli. 2025. "Genotype Analysis on Orientia tsutsugamushi Causing Scrub Typhus in Malaysia: A Re-Emerging Disease" Tropical Medicine and Infectious Disease 10, no. 9: 252. https://doi.org/10.3390/tropicalmed10090252
APA StyleHii, S. Y. F., Mohd Zaidi, M. N., Kassim, W. N., Hashim, R., & Ramli, S. R. (2025). Genotype Analysis on Orientia tsutsugamushi Causing Scrub Typhus in Malaysia: A Re-Emerging Disease. Tropical Medicine and Infectious Disease, 10(9), 252. https://doi.org/10.3390/tropicalmed10090252





