Building Pathogen Genomic Sequencing Capacity in Africa: Centre for Epidemic Response and Innovation Fellowship
Abstract
1. Introduction
2. Programme Description
2.1. Selection of Fellows
2.2. Learning Objectives of the Training
- Hands-on training in wet-bench protocols, including nucleic acid amplification, library preparation, and sequencing.
- Adaptation of protocols to sequencing other viral and bacterial pathogens.
- Pathogen genomic bioinformatics, which included an introduction to foundational computational skills, base-calling and sequence assembly, sequence quality control checks, and submission of sequence data to public genomic repositories.
- Basic phylogenetic analysis, which included compiling a sequence alignment and building phylogenetic trees.
- Reporting sequencing results and communicating scientific findings for public health applications.
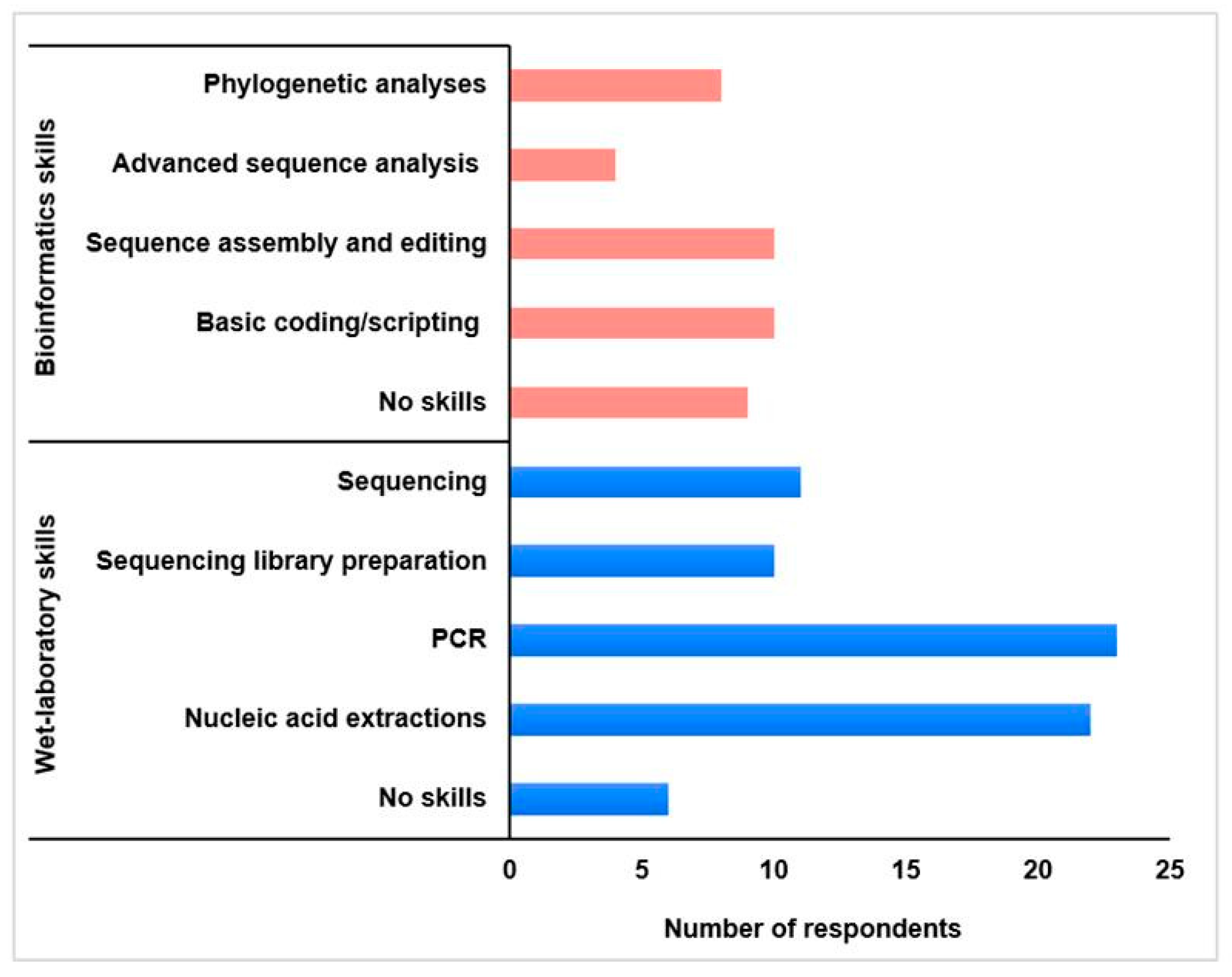
2.3. Fellows’ Professional Profiles
2.4. The Training Event
2.5. Approaches to Gather Fellows’ Perspectives
3. Discussion
3.1. Fellows’ Perspectives on the Training Outcomes
3.1.1. Acquisition of Specialised Skills and Knowledge Advancement
3.1.2. Revolution of Next-Generation Sequencing (NGS)
3.1.3. Genomic Surveillance of Pathogens: Real-Time Detection and Results Communication
3.1.4. Protocol Adaptation and Miniaturisation
3.1.5. In-Person Networking and Collaboration
3.2. Critical Evaluation of the Training Programme
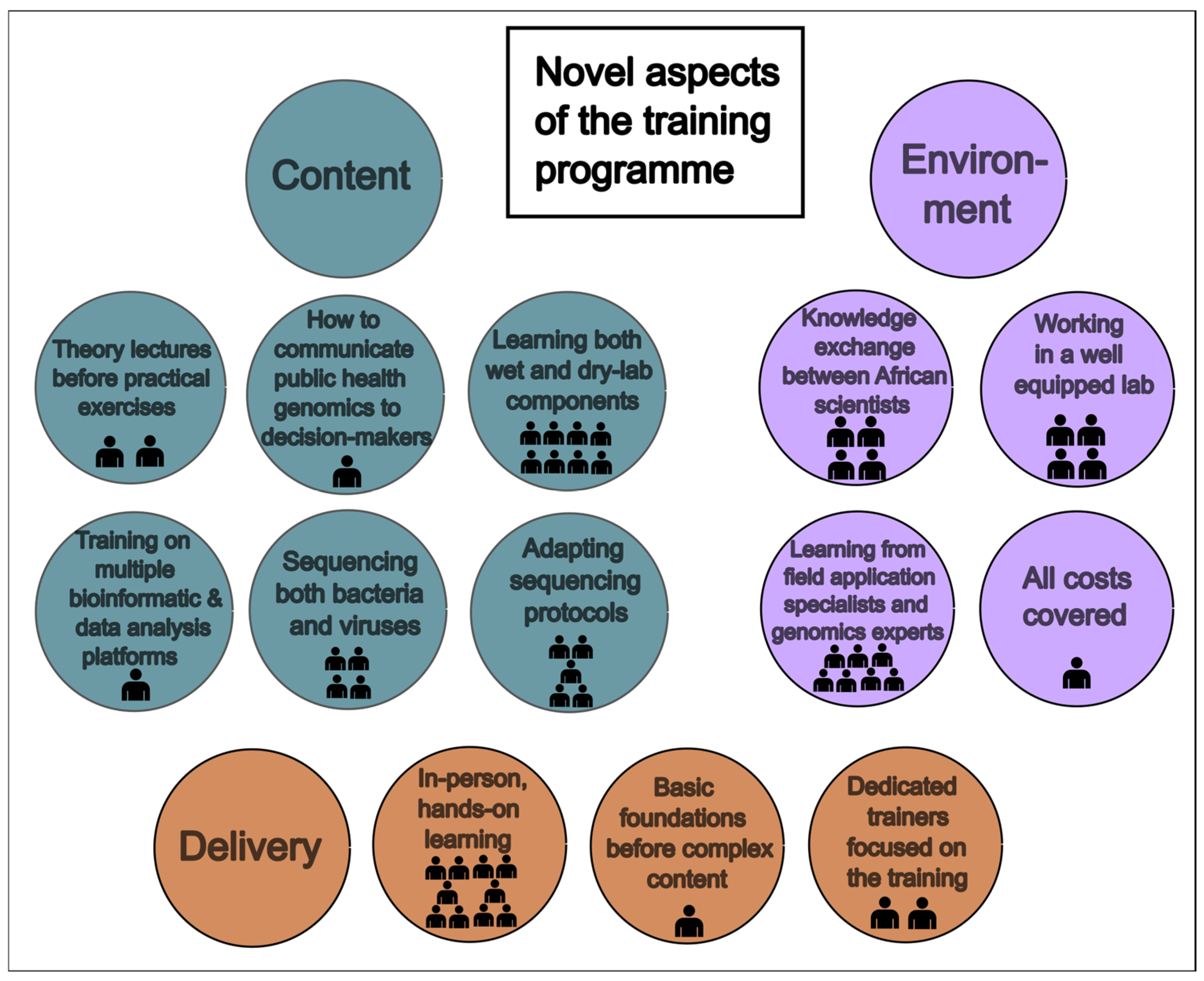
3.2.1. Novel Approaches
3.2.2. Train-the-Trainer
3.2.3. Limitations and Suggestions for Improvement
3.3. Fellows’ Knowledge Transfer at Home Institutes
3.3.1. Knowledge and Skills Sharing
3.3.2. Implementation of Learnt Skills and Knowledge Transfer by Dr. Molalegne Bitew
3.4. The Way Forward for Africa: Better Together
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fatumo, S.; Chikowore, T.; Choudhury, A.; Ayub, M.; Martin, A.R.; Kuchenbaecker, K. A Roadmap to Increase Diversity in Genomic Studies. Nat. Med. 2022, 28, 243–250. [Google Scholar]
- Chabuka, L.; Choga, W.T.; Mavian, C.N.; Moir, M.; Tegally, H.; Wilkinson, E.; Naidoo, Y.; Inward, R.; Morgenstern, C.; Bhatt, S.; et al. Genomic Epidemiology of the Cholera Outbreak in Malawi 2022–2023. medRxiv 2023. medRxiv:2023.08.22.23294324. [Google Scholar] [CrossRef]
- Jackson, B.R.; Tarr, C.; Strain, E.; Jackson, K.A.; Conrad, A.; Carleton, H.; Katz, L.S.; Stroika, S.; Gould, L.H.; Mody, R.K.; et al. Implementation of Nationwide Real-Time Whole-Genome Sequencing to Enhance Listeriosis Outbreak Detection and Investigation. Clin. Infect. Dis. 2016, 63, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Oude Munnink, B.B.; Nieuwenhuijse, D.F.; Stein, M.; O’Toole, Á.; Haverkate, M.; Mollers, M.; Kamga, S.K.; Schapendonk, C.; Pronk, M.; Lexmond, P.; et al. Rapid SARS-CoV-2 Whole-Genome Sequencing and Analysis for Informed Public Health Decision-Making in the Netherlands. Nat. Med. 2020, 26, 1405–1410. [Google Scholar] [PubMed]
- Tastan Bishop, O.; Adebiyi, E.F.; Alzohairy, A.M.; Everett, D.; Ghedira, K.; Ghouila, A.; Kumuthini, J.; Mulder, N.J.; Panji, S.; Patterton, H.-G. Bioinformatics Education--Perspectives and Challenges out of Africa. Brief. Bioinform. 2015, 16, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Brito, A.F.; Semenova, E.; Dudas, G.; Hassler, G.W.; Kalinich, C.C.; Kraemer, M.U.G.; Ho, J.; Tegally, H.; Githinji, G.; Agoti, C.N.; et al. Global Disparities in SARS-CoV-2 Genomic Surveillance. Nat. Commun. 2022, 13, 7003. [Google Scholar]
- Tekola-Ayele, F.; Rotimi, C.N. Translational Genomics in Low- and Middle-Income Countries: Opportunities and Challenges. Public Health Genom. 2015, 18, 242–247. [Google Scholar]
- Tegally, H.; San, J.E.; Cotten, M.; Moir, M.; Tegomoh, B.; Mboowa, G.; Martin, D.P.; Baxter, C.; Lambisia, A.W.; Diallo, A.; et al. The Evolving SARS-CoV-2 Epidemic in Africa: Insights from Rapidly Expanding Genomic Surveillance. Science 2022, 378, eabq5358. [Google Scholar]
- de Oliveira, T.; Baxter, C. Investing in Africa’s Scientific Future. Science 2024, 383, eadn4168. [Google Scholar]
- Rugarabamu, S.; Mboera, L.; Rweyemamu, M.; Mwanyika, G.; Lutwama, J.; Paweska, J.; Misinzo, G. Forty-Two Years of Responding to Ebola Virus Outbreaks in Sub-Saharan Africa: A Review. BMJ Glob. Health 2020, 5, e001955. [Google Scholar]
- Lowe, R.; Barcellos, C.; Brasil, P.; Cruz, O.; Honório, N.; Kuper, H.; Carvalho, M. The Zika Virus Epidemic in Brazil: From Discovery to Future Implications. Int. J. Environ. Res. Public Health 2018, 15, 96. [Google Scholar] [CrossRef] [PubMed]
- QGIS. QGIS Geographic Information System; Open-Source Geospatial Foundation Projects; OSGeo: Beaverton, OR, USA, 2023. [Google Scholar]
- Pillay, S.; San, J.E.; Tshiabuila, D.; Naidoo, Y.; Pillay, Y.; Maharaj, A.; Anyaneji, U.J.; Wilkinson, E.; Tegally, H.; Lessells, R.J.; et al. Evaluation of Miniaturized Illumina DNA Preparation Protocols for SARS-CoV-2 Whole Genome Sequencing. PLoS ONE 2023, 18, e0283219. [Google Scholar] [CrossRef]
- Stassen, W. Institute Leads Cutting-Edge Biomedical Research in Africa, for Africa. Available online: https://www0.sun.ac.za/researchforimpact/2023/08/29/institute-leads-cutting-edge-biomedical-research-in-africa-for-africa/#:~:text=A world-class biomedical research,its facilities and research capacity (accessed on 12 January 2024).
- Koboldt, D.C.; Steinberg, K.M.; Larson, D.E.; Wilson, R.K.; Mardis, E.R. The Next-Generation Sequencing Revolution and Its Impact on Genomics. Cell 2013, 155, 27–38. [Google Scholar] [CrossRef]
- Adebisi, Y.A.; Rabe, A.; Lucero-Prisno, D.E., III. COVID-19 Surveillance Systems in African Countries. Health Promot. Perspect. 2021, 11, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Xavier, J.S.; Moir, M.; Tegally, H.; Sitharam, N.; Abdool Karim, W.; San, J.E.; Linhares, J.; Wilkinson, E.; Ascher, D.B.; Baxter, C.; et al. SARS-CoV-2 Africa Dashboard for Real-Time COVID-19 Information. Nat. Microbiol. 2022, 8, 1–4. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global Initiative on Sharing All Influenza Data—From Vision to Reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef] [PubMed]
- Kabir, F.; Plaisance, E.; Portman, A.; Marfo, A.; Cirrincione, K.; Silva, D.; Amadi, V.; Stringer, J.; Short, L. Mpox Viral Lineage Analysis and Technique Development Using Next-Generation Sequencing Approach. J. Infect. Dis. 2024, 229, S163–S171. [Google Scholar] [CrossRef] [PubMed]
- Davina-Nunez, C.; Perez-Castro, S.; Cabrera-Alvargonzalez, J.J.; Montano-Barrientos, J.; Godoy-Diz, M.; Regueiro, B. The Modification of the Illumina® CovidSeqTM Workflow for RSV Genomic Surveillance: The Genetic Variability of RSV during the 2022–2023 Season in Northwest Spain. Int. J. Mol. Sci. 2023, 24, 16055. [Google Scholar] [CrossRef]
- Pillay, S.; Giandhari, J.; Tegally, H.; Wilkinson, E.; Chimukangara, B.; Lessells, R.; Moosa, Y.; Mattison, S.; Gazy, I.; Fish, M.; et al. Whole Genome Sequencing of SARS-CoV-2: Adapting Illumina Protocols for Quick and Accurate Outbreak Investigation during a Pandemic. Genes 2020, 11, 949. [Google Scholar] [CrossRef]
- Hailu, G.; Legesse, M.; Mulu, A.; Medhin, G.; Mengesha, M.; Hailu, D.; Ayele, A.; Gebreegziabxier, A.; Tayachew, A.; Aguine, A.; et al. SARS-CoV-2 Genetic Variants Identified in Selected Regions of Ethiopia through Whole Genome Sequencing: Insights from the Fifth Wave of COVID-19. Genes 2025, 16, 351. [Google Scholar] [CrossRef] [PubMed]

| Training Topics | Time Allocated (h) |
|---|---|
| Wet-Laboratory Training Components | |
| DNA/RNA amplification | 6.5 |
| Library preparation | 19.5 |
| Sequencing | 8 |
| Adapting protocols to other pathogens | 3 |
| Illumina sequencing technology | 2.5 |
| Recap and Q&A | 3 |
| Bioinformatics Training Components | |
| Introduction to Unix | 1.5 |
| Introduction to bash scripting | 1.5 |
| Introduction to R and RStudio | 2 |
| Base-calling and assembly | 1.5 |
| Sequence assembly and analysis | 6 |
| Sequence quality control | 2 |
| Sequence submission to public databases | 1.5 |
| Basic phylogenetics | 5 |
| Recap and Q&A | 3 |
| Science Communication for Public Health | 1.5 |
| Scientific Seminars | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agboli, E.; Bitew, M.; Malaka, C.N.; Kallon, T.M.P.S.; Jalloh, A.M.S.; Yankonde, B.; Shempela, D.M.; Sikalima, J.F.M.; Joseph, M.; Kasonde, M.; et al. Building Pathogen Genomic Sequencing Capacity in Africa: Centre for Epidemic Response and Innovation Fellowship. Trop. Med. Infect. Dis. 2025, 10, 90. https://doi.org/10.3390/tropicalmed10040090
Agboli E, Bitew M, Malaka CN, Kallon TMPS, Jalloh AMS, Yankonde B, Shempela DM, Sikalima JFM, Joseph M, Kasonde M, et al. Building Pathogen Genomic Sequencing Capacity in Africa: Centre for Epidemic Response and Innovation Fellowship. Tropical Medicine and Infectious Disease. 2025; 10(4):90. https://doi.org/10.3390/tropicalmed10040090
Chicago/Turabian StyleAgboli, Eric, Molalegne Bitew, Christian N. Malaka, Tiangay M. P. S. Kallon, Alhaji M. S. Jalloh, Baron Yankonde, Doreen M. Shempela, Jay F. M. Sikalima, Mutale Joseph, Mpanga Kasonde, and et al. 2025. "Building Pathogen Genomic Sequencing Capacity in Africa: Centre for Epidemic Response and Innovation Fellowship" Tropical Medicine and Infectious Disease 10, no. 4: 90. https://doi.org/10.3390/tropicalmed10040090
APA StyleAgboli, E., Bitew, M., Malaka, C. N., Kallon, T. M. P. S., Jalloh, A. M. S., Yankonde, B., Shempela, D. M., Sikalima, J. F. M., Joseph, M., Kasonde, M., Demeke, F. M., Valdese, A. F. I., Grace, L. B., Célestin, G., Papkiauri, A., Berlange, S. Y. F., Majanja, J., Omwenga, V. K., Wambugu, E. N., ... Moir, M. (2025). Building Pathogen Genomic Sequencing Capacity in Africa: Centre for Epidemic Response and Innovation Fellowship. Tropical Medicine and Infectious Disease, 10(4), 90. https://doi.org/10.3390/tropicalmed10040090







