Molecular Characterization of Plasmodium Species to Strengthen Malaria Surveillance in Migrant Populations in Honduras
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Molecular Diagnosis
2.3. Molecular Markers for P. falciparum: pfcrt, pfmdr1, pfglurp, pfama1, pfhrp2/3, and pfs47
2.4. Molecular Markers for P. vivax: pvcsp, pvmsp1-F2 block, pvmsp3α, pvmsp3β, pvs47, and pvs48/45
3. Results
3.1. Molecular Markers for P. falciparum
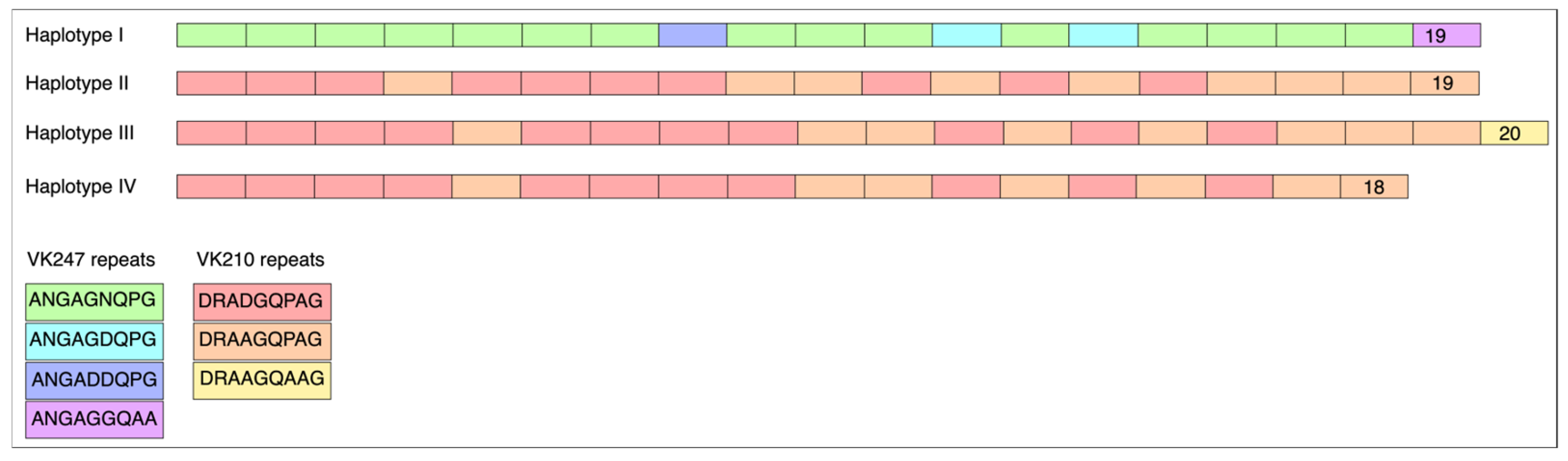
3.2. Molecular Markers for P. vivax
pvs47 and pvs48/45
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NGS | Next Generation Sequencing |
| CQ | Chloroquine |
| Pvcsp | Plasmodium vivax circumsporozoite protein |
| Pvmsp3a | Plasmodium vivax merozoite surface protein 2 alpha |
| Pvmsp3b | Plasmodium vivax merozoite surface protein 2 beta |
| Pvmsp1 | Plasmodium vivax merozoite surface protein 1 |
| Pvs47 | Plasmodium vivax surface protein 47 |
| Pvs48/45 | Plasmodium vivax surface protein 48/45 |
| Pfcrt | Plasmodium falciparum chloroquine transporter |
| Pfmdr1 | Plasmodium falciparum multidrug resistance 1 |
| Pfhrp2/3 | Plasmodium falciparum histidine rich proteins 2/3 |
| Pfama1 | Plasmodium falciparum apical membrane antigen 1 |
| Pfs47 | Plasmodium falciparum surface protein 47 |
References
- World Health Organization. World Malaria Report 2024; Global Malaria Programme (GMP), WHO: Geneva, Switzerland, 2024; p. 316. [Google Scholar]
- Pinto, A.; Archaga, O.; Mejia, A.; Escober, L.; Henriquez, J.; Montoya, A.; Valdivia, H.O.; Fontecha, G. Evidence of a Recent Bottleneck in Plasmodium falciparum Populations on the Honduran-Nicaraguan Border. Pathogens 2021, 10, 1432. [Google Scholar] [CrossRef]
- Zamora, A.; Pinto, A.; Escobar, D.; Valdivia, H.O.; Chaver, L.; Ardon, G.; Carranza, E.; Fontecha, G. Genetic diversity of Plasmodium vivax and Plasmodium falciparum field isolates from Honduras in the malaria elimination phase. Curr. Res. Parasitol. Vector Borne Dis. 2025, 7, 100230. [Google Scholar] [CrossRef]
- Agudelo Higuita, N.I.; Franco-Paredes, C.; Henao-Martinez, A.F.; Mendez Rojas, B.; Suarez, J.A.; Naranjo, L.; Alger, J. Migrants in transit across Central America and the potential spread of chloroquine resistant malaria-a call for action. Lancet Reg. Health Am. 2023, 22, 100505. [Google Scholar] [CrossRef]
- Loyola-Cruz, M.A.; Duran-Manuel, E.M.; Cruz-Cruz, C.; Bravata-Alcantara, J.C.; Gutierrez-Munoz, V.H.; Marquez-Valdelamar, L.M.; Leal-Escobar, B.; Vasquez-Jimenez, E.; Cureno-Diaz, M.A.; Lugo-Zamudio, G.E.; et al. Imported malaria cases by Plasmodium falciparum and Plasmodium vivax in Mexican territory: Potential impact of the migration crisis. Travel Med. Infect. Dis. 2024, 62, 102773. [Google Scholar] [CrossRef]
- Fontecha, G. The Honduran diaspora and infectious diseases: An urgent need for action. Travel Med. Infect. Dis. 2023, 53, 102567. [Google Scholar] [CrossRef]
- He, X.; Zhong, D.; Zou, C.; Pi, L.; Zhao, L.; Qin, Y.; Pan, M.; Wang, S.; Zeng, W.; Xiang, Z.; et al. Unraveling the Complexity of Imported Malaria Infections by Amplicon Deep Sequencing. Front. Cell. Infect. Microbiol. 2021, 11, 725859. [Google Scholar] [CrossRef]
- Secretaría de Salud de Honduras. Norma Nacional de Malaria de Honduras; Secretaría de Salud de Honduras: Tegucigalpa, Honduras, 2018. [Google Scholar]
- Balikagala, B.; Sakurai-Yatsushiro, M.; Tachibana, S.I.; Ikeda, M.; Yamauchi, M.; Katuro, O.T.; Ntege, E.H.; Sekihara, M.; Fukuda, N.; Takahashi, N.; et al. Recovery and stable persistence of chloroquine sensitivity in Plasmodium falciparum parasites after its discontinued use in Northern Uganda. Malar. J. 2020, 19, 76. [Google Scholar] [CrossRef]
- Dakorah, M.P.; Aninagyei, E.; Attoh, J.; Adzakpah, G.; Tukwarlba, I.; Acheampong, D.O. Profiling antimalarial drug-resistant haplotypes in Pfcrt, Pfmdr1, Pfdhps and Pfdhfr genes in Plasmodium falciparum causing malaria in the Central Region of Ghana: A multicentre cross-sectional study. Ther. Adv. Infect. Dis. 2025, 12, 20499361251319665. [Google Scholar] [CrossRef]
- Sathishkumar, V.; Nirmolia, T.; Bhattacharyya, D.R.; Patgiri, S.J. Genetic polymorphism of Plasmodium falciparum msp-1, msp-2 and glurp vaccine candidate genes in pre-artemisinin era clinical isolates from Lakhimpur district in Assam, Northeast India. Access Microbiol. 2022, 4, 000350. [Google Scholar] [CrossRef]
- Thanapongpichat, S.; Khammanee, T.; Sawangjaroen, N.; Buncherd, H.; Tun, A.W. Genetic Diversity of Plasmodium vivax in Clinical Isolates from Southern Thailand using PvMSP1, PvMSP3 (PvMSP3alpha, PvMSP3beta) Genes and Eight Microsatellite Markers. Korean J. Parasitol. 2019, 57, 469–479. [Google Scholar] [CrossRef]
- Ciubotariu, I.I.; Broyles, B.K.; Xie, S.; Thimmapuram, J.; Mwenda, M.C.; Mambwe, B.; Mulube, C.; Matoba, J.; Schue, J.L.; Moss, W.J.; et al. Diversity and selection analyses identify transmission-blocking antigens as the optimal vaccine candidates in Plasmodium falciparum. medRxiv 2024. [Google Scholar] [CrossRef]
- Barillas-Mury, C.; Molina-Cruz, A.; Torres, T.; Raytselis, N.; Young, M.; Kamath, N.; McNich, C.; Su, X.-Z.; Dwivedi, A.; Silva, J.; et al. Worldwide genetic diversity and population structure of Plasmodium vivax Pv47 is consistent with natural selection by anopheline mosquitoes. Res. Sq. 2024. [Google Scholar] [CrossRef]
- Fontecha, G.; Escobar, D.; Ortiz, B.; Pinto, A. A PCR-RFLP Technique to Assess the Geographic Origin of Plasmodium falciparum Strains in Central America. Trop. Med. Infect. Dis. 2022, 7, 149. [Google Scholar] [CrossRef]
- Woo, M.K.; Kim, K.A.; Kim, J.; Oh, J.S.; Han, E.T.; An, S.S.; Lim, C.S. Sequence polymorphisms in Pvs48/45 and Pvs47 gametocyte and gamete surface proteins in Plasmodium vivax isolated in Korea. Mem. Inst. Oswaldo Cruz 2013, 108, 359–367. [Google Scholar] [CrossRef]
- Valdivia, H.O.; Villena, F.E.; Lizewski, S.E.; Garcia, J.; Alger, J.; Bishop, D.K. Genomic surveillance of Plasmodium falciparum and Plasmodium vivax cases at the University Hospital in Tegucigalpa, Honduras. Sci. Rep. 2020, 10, 20975. [Google Scholar] [CrossRef]
- Abdallah, J.F.; Okoth, S.A.; Fontecha, G.A.; Torres, R.E.; Banegas, E.I.; Matute, M.L.; Bucheli, S.T.; Goldman, I.F.; de Oliveira, A.M.; Barnwell, J.W.; et al. Prevalence of pfhrp2 and pfhrp3 gene deletions in Puerto Lempira, Honduras. Malar. J. 2015, 14, 19. [Google Scholar] [CrossRef]
- Fontecha, G.; Mejia, R.E.; Banegas, E.; Ade, M.P.; Mendoza, L.; Ortiz, B.; Sabillon, I.; Alvarado, G.; Matamoros, G.; Pinto, A. Deletions of pfhrp2 and pfhrp3 genes of Plasmodium falciparum from Honduras, Guatemala and Nicaragua. Malar. J. 2018, 17, 320. [Google Scholar] [CrossRef]
- Fontecha, G.; Pinto, A.; Archaga, O.; Betancourth, S.; Escober, L.; Henriquez, J.; Valdivia, H.O.; Montoya, A.; Mejia, R.E. Assessment of Plasmodium falciparum anti-malarial drug resistance markers in pfcrt and pfmdr1 genes in isolates from Honduras and Nicaragua, 2018–2021. Malar. J. 2021, 20, 465. [Google Scholar] [CrossRef]
- Fontecha, G.A.; Sanchez, A.L.; Mendoza, M.; Banegas, E.; Mejia-Torres, R.E. A four-year surveillance program for detection of Plasmodium falciparum chloroquine resistance in Honduras. Mem. Inst. Oswaldo Cruz 2014, 109, 492–493. [Google Scholar] [CrossRef]
- Jovel, I.T.; Mejia, R.E.; Banegas, E.; Piedade, R.; Alger, J.; Fontecha, G.; Ferreira, P.E.; Veiga, M.I.; Enamorado, I.G.; Bjorkman, A.; et al. Drug resistance associated genetic polymorphisms in Plasmodium falciparum and Plasmodium vivax collected in Honduras, Central America. Malar. J. 2011, 10, 376. [Google Scholar] [CrossRef]
- Lefebvre, M.J.M.; Degrugillier, F.; Arnathau, C.; Fontecha, G.A.; Noya, O.; Houze, S.; Severini, C.; Pradines, B.; Berry, A.; Trape, J.F.; et al. Genomic exploration of the journey of Plasmodium vivax in Latin America. PLoS Pathog. 2025, 21, e1012811. [Google Scholar] [CrossRef]
- Lopez, A.C.; Ortiz, A.; Coello, J.; Sosa-Ochoa, W.; Torres, R.E.; Banegas, E.I.; Jovel, I.; Fontecha, G.A. Genetic diversity of Plasmodium vivax and Plasmodium falciparum in Honduras. Malar. J. 2012, 11, 391. [Google Scholar] [CrossRef]
- Matamoros, G.; Escobar, D.; Pinto, A.; Serrano, D.; Ksandrova, E.; Grimaldi, N.; Juarez-Fontecha, G.; Moncada, M.; Valdivia, H.O.; Fontecha, G. PET-PCR reveals low parasitaemia and submicroscopic malarial infections in Honduran Moskitia. Malar. J. 2023, 22, 110. [Google Scholar] [CrossRef]
- Ortiz-Morazan, A.S.; Moncada, M.M.; Escobar, D.; Cabrera-Moreno, L.A.; Fontecha, G. Coevolutionary Analysis of the Pfs47-P47Rec Complex: A Bioinformatics Approach. Bioinform. Biol. Insights 2024, 18, 11779322241284223. [Google Scholar] [CrossRef]
- Lucchi, N.W.; Narayanan, J.; Karell, M.A.; Xayavong, M.; Kariuki, S.; DaSilva, A.J.; Hill, V.; Udhayakumar, V. Molecular diagnosis of malaria by photo-induced electron transfer fluorogenic primers: PET-PCR. PLoS ONE 2013, 8, e56677. [Google Scholar] [CrossRef]
- Kudyba, H.M.; Louzada, J.; Ljolje, D.; Kudyba, K.A.; Muralidharan, V.; Oliveira-Ferreira, J.; Lucchi, N.W. Field evaluation of malaria malachite green loop-mediated isothermal amplification in health posts in Roraima state, Brazil. Malar. J. 2019, 18, 98. [Google Scholar] [CrossRef]
- Griffing, S.; Syphard, L.; Sridaran, S.; McCollum, A.M.; Mixson-Hayden, T.; Vinayak, S.; Villegas, L.; Barnwell, J.W.; Escalante, A.A.; Udhayakumar, V. pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob. Agents Chemother. 2010, 54, 1572–1579. [Google Scholar] [CrossRef]
- Kang, J.M.; Lee, J.; Moe, M.; Jun, H.; Le, H.G.; Kim, T.I.; Thai, T.L.; Sohn, W.M.; Myint, M.K.; Lin, K.; et al. Population genetic structure and natural selection of Plasmodium falciparum apical membrane antigen-1 in Myanmar isolates. Malar. J. 2018, 17, 71. [Google Scholar] [CrossRef]
- Molina-Cruz, A.; Raytselis, N.; Withers, R.; Dwivedi, A.; Crompton, P.D.; Traore, B.; Carpi, G.; Silva, J.C.; Barillas-Mury, C. A genotyping assay to determine geographic origin and transmission potential of Plasmodium falciparum malaria cases. Commun. Biol. 2021, 4, 1145. [Google Scholar] [CrossRef]
- Imwong, M.; Pukrittayakamee, S.; Gruner, A.C.; Renia, L.; Letourneur, F.; Looareesuwan, S.; White, N.J.; Snounou, G. Practical PCR genotyping protocols for Plasmodium vivax using Pvcs and Pvmsp1. Malar. J. 2005, 4, 20. [Google Scholar] [CrossRef]
- Han, E.T.; Park, J.H.; Shin, E.H.; Choi, M.H.; Oh, M.D.; Chai, J.Y. Apical membrane antigen-1 (AMA-1) gene sequences of re-emerging Plasmodium vivax in South Korea. Korean J. Parasitol. 2002, 40, 157–162. [Google Scholar] [CrossRef]
- Tachibana, M.; Suwanabun, N.; Kaneko, O.; Iriko, H.; Otsuki, H.; Sattabongkot, J.; Kaneko, A.; Herrera, S.; Torii, M.; Tsuboi, T. Plasmodium vivax gametocyte proteins, Pvs48/45 and Pvs47, induce transmission-reducing antibodies by DNA immunization. Vaccine 2015, 33, 1901–1908. [Google Scholar] [CrossRef]
- Vallejo, A.F.; Martinez, N.L.; Tobon, A.; Alger, J.; Lacerda, M.V.; Kajava, A.V.; Arevalo-Herrera, M.; Herrera, S. Global genetic diversity of the Plasmodium vivax transmission-blocking vaccine candidate Pvs48/45. Malar. J. 2016, 15, 202. [Google Scholar] [CrossRef]
- Cao, Y.; Hart, R.J.; Bansal, G.P.; Kumar, N. Functional Conservation of P48/45 Proteins in the Transmission Stages of Plasmodium vivax (Human Malaria Parasite) and P. berghei (Murine Malaria Parasite). mBio 2018, 9, e01627-18. [Google Scholar] [CrossRef]
- Kang, J.M.; Le, H.G.; Vo, T.C.; Naw, H.; Yoo, W.G.; Sohn, W.M.; Trinh, N.T.M.; Quang, H.H.; Na, B.K. Genetic Polymorphism and Natural Selection of Apical Membrane Antigen-1 in Plasmodium falciparum Isolates from Vietnam. Genes 2021, 12, 1903. [Google Scholar] [CrossRef]
- Tatem, A.J.; Jia, P.; Ordanovich, D.; Falkner, M.; Huang, Z.; Howes, R.; Hay, S.I.; Gething, P.W.; Smith, D.L. The geography of imported malaria to non-endemic countries: A meta-analysis of nationally reported statistics. Lancet Infect. Dis. 2017, 17, 98–107. [Google Scholar] [CrossRef]
- Oyegoke, O.O.; Maharaj, L.; Akoniyon, O.P.; Kwoji, I.; Roux, A.T.; Adewumi, T.S.; Maharaj, R.; Oyebola, B.T.; Adeleke, M.A.; Okpeku, M. Malaria diagnostic methods with the elimination goal in view. Parasitol. Res. 2022, 121, 1867–1885. [Google Scholar] [CrossRef]
- Banegas, S.; Escobar, D.; Pinto, A.; Moncada, M.; Matamoros, G.; Valdivia, H.O.; Reyes, A.; Fontecha, G. Asymptomatic Malaria Reservoirs in Honduras: A Challenge for Elimination. Pathogens 2024, 13, 541. [Google Scholar] [CrossRef]
- Pierre-Louis, E.; Kelley, J.; Patel, D.; Carlson, C.; Talundzic, E.; Jacobson, D.; Barratt, J.L.N. Geo-classification of drug-resistant travel-associated Plasmodium falciparum using Pfs47 and Pfcpmp gene sequences (USA, 2018–2021). Antimicrob. Agents Chemother. 2024, 68, e0120324. [Google Scholar] [CrossRef]
- Takahashi, N.; Tanabe, K.; Tsukahara, T.; Dzodzomenyo, M.; Dysoley, L.; Khamlome, B.; Sattabongkot, J.; Nakamura, M.; Sakurai, M.; Kobayashi, J.; et al. Large-scale survey for novel genotypes of Plasmodium falciparum chloroquine-resistance gene pfcrt. Malar. J. 2012, 11, 92. [Google Scholar] [CrossRef]
- Mejia Torres, R.E.; Banegas, E.I.; Mendoza, M.; Diaz, C.; Bucheli, S.T.; Fontecha, G.A.; Alam, M.T.; Goldman, I.; Udhayakumar, V.; Zambrano, J.O. Efficacy of chloroquine for the treatment of uncomplicated Plasmodium falciparum malaria in Honduras. Am. J. Trop. Med. Hyg. 2013, 88, 850–854. [Google Scholar] [CrossRef]
- Mohammed, A.; Ndaro, A.; Kalinga, A.; Manjurano, A.; Mosha, J.F.; Mosha, D.F.; van Zwetselaar, M.; Koenderink, J.B.; Mosha, F.W.; Alifrangis, M.; et al. Trends in chloroquine resistance marker, Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania. Malar. J. 2013, 12, 415. [Google Scholar] [CrossRef]
- Musyoka, K.B.; Kiiru, J.N.; Aluvaala, E.; Omondi, P.; Chege, W.K.; Judah, T.; Kiboi, D.; Nganga, J.K.; Kimani, F.T. Prevalence of mutations in Plasmodium falciparum genes associated with resistance to different antimalarial drugs in Nyando, Kisumu County in Kenya. Infect. Genet. Evol. 2020, 78, 104121. [Google Scholar] [CrossRef]
- Sarah-Matio, E.M.; Guillochon, E.; Nsango, S.E.; Abate, L.; Ngou, C.M.; Bouopda, G.A.; Feufack-Donfack, L.B.; Bayibeki, A.N.; Tchioffo Tsapi, M.; Talman, A.; et al. Genetic Diversity of Plasmodium falciparum and Distribution of Antimalarial Drug Resistance Mutations in Symptomatic and Asymptomatic Infections. Antimicrob. Agents Chemother. 2022, 66, e0018822. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, X.; Chen, H.; Zhang, J.; Lu, Q.; Ruan, W.; Wang, X. Molecular markers associated with drug resistance in Plasmodium falciparum parasites in central Africa between 2016 and 2021. Front. Public Health 2023, 11, 1239274. [Google Scholar] [CrossRef]
- Zhang, J.J.; Senaratne, T.N.; Daniels, R.; Valim, C.; Alifrangis, M.; Amerasinghe, P.; Konradsen, F.; Rajakaruna, R.; Wirth, D.F.; Karunaweera, N.D. Distribution pattern of Plasmodium falciparum chloroquine transporter (pfcrt) gene haplotypes in Sri Lanka 1996–2006. Am. J. Trop. Med. Hyg. 2011, 85, 811–814. [Google Scholar] [CrossRef]
- Gamboa, D.; Ho, M.F.; Bendezu, J.; Torres, K.; Chiodini, P.L.; Barnwell, J.W.; Incardona, S.; Perkins, M.; Bell, D.; McCarthy, J.; et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: Implications for malaria rapid diagnostic tests. PLoS ONE 2010, 5, e8091. [Google Scholar] [CrossRef]
- Molina-Cruz, A.; Canepa, G.E.; Alves, E.S.T.L.; Williams, A.E.; Nagyal, S.; Yenkoidiok-Douti, L.; Nagata, B.M.; Calvo, E.; Andersen, J.; Boulanger, M.J.; et al. Plasmodium falciparum evades immunity of anopheline mosquitoes by interacting with a Pfs47 midgut receptor. Proc. Natl. Acad. Sci. USA 2020, 117, 2597–2605. [Google Scholar] [CrossRef]
| Target Gene | Reaction | Primer Name | Primer Sequence (5′-3′) | References |
|---|---|---|---|---|
| 18Sr RNA gene | PET-PCR for genus Plasmodium | Genus forward | GGC CTA ACA TGG CTA TGA CG | [25,27] |
| Genus reverse | 6FAM-agg cgc ata gcg cct gg CTG CCT TCC TTA GAT GTG GTA GCT | |||
| 18Sr RNA gene | PET-PCR for P. falciparum | Falciparum forward | ACC CCT CGC CTG GTG TTT TT | [27] |
| Falciparum reverse | HEX-agg cgc ata gcg cct gg TCG GGC CCC AAA AAT AGG AA | |||
| 18Sr RNA gene | PET-PCR for P. vivax | Vivax forward | ACT GAC ACT GAT GAT TTA GAA CCC ATT T | [28] |
| Vivax reverse | HEX- agg cgc ata gcg cct ggT GGA GAG ATC TTT CCA TCC TAA ACC T | |||
| pfcrt | 1st round | AL6821 | AGC AAA AAT GAC GAG CGT TAT AG | [3,29] |
| AL6822 | ATT GGT AGG TGG AAT AGA TTC TC | |||
| 2nd round | AL5631 | TTT TTC CCT TGT CGA CCT TAA C | ||
| AL5632 | AGG AAT AAA CAA TAA AGA ACA TAA TCA TAC | |||
| pfmdr1 | SNPs 86, 184 | MDR1-1F | TTA AAT GTT TAC CTG CAC AAC ATA GAA AAT T | [20] |
| MDR1-1R | CTC CAC AAT AAC TTG CAA CAG TTC TTA | |||
| MDR1-2F | TGT ATG TGC TGT ATT ATC AGGA | |||
| MDR1-2R | CTC TTC TAT AAT GGA CAT GGTA | |||
| pfmdr1 | SNPs 1034, 1046 | 1042-A | GTC GAA AAG ACT ATG AAA CGT AGA | [20] |
| 1042-C | CTC AAA TGA TAA TTT TGC AT | |||
| 1042-B | GAT CCA AGT TTT TTA ATA CA | |||
| 1042-C | CTC AAA TGA TAA TTT TGC AT | |||
| pfmdr1 | SNP 1246 | 1246-A | GTG GAA AAT CAA CTT TTA TGA | [20] |
| 1246-B | TTA GGT TCT CTT AAT AAT GCT | |||
| 1246-C | GAC TTG AAA AAT GAT CAC ATT | |||
| 1246-D | GTC CAC CTG ATA TGC TTT T | |||
| pfglurp | 1st round | GLURPOF | TGA ATT TGA AGA TGT TCA CAC TGA AC | [3,11] |
| GLURPOR | GTG GAA TTG CTT TTT CTT CAA CAC TAA | |||
| 2nd round | GLURPGNF | TGT TCA CAC TGA ACA ATT AGA TTT AGA TCA | ||
| GLURPOR | GTG GAA TTG CTT TTT CTT CAA CAC TAA | |||
| pfama1 | 1st round | Pfama1F | GTA CTT GTT ATA AAT TGT ACA | [30] |
| Pfama1R | TTT TAG CAT AAA AGA GAA GC | |||
| 2nd round | Pfama1F1 | ACA AAA ATG AGA AAA TTA TAC TGC | [30] | |
| Pfama1R1 | TTA ATA GTA TGG TTT TTC CAT CAG AAC | |||
| pfhrp2 exons 1–2 | 1st round | 2E12F1 | GGT TTC CTT CTC AAA AAA TAA AG | [18,19] |
| 2E12R1 | TCT ACA TGT GCT TGA GTT TCG | |||
| 2nd round | 2E12F | GTA TTA TCC GCT GCC GTT TTT GCC | ||
| 2E12R | CTA CAC AAG TTA TTA TTA AAT GCG GAA | |||
| pfhrp3 exons 1–2 | 1st round | 3E12F1 | GGT TTC CTT CTC AAA AAA TAA AA | [18,19] |
| 3E12R1 | CCT GCA TGT GCT TGA CTT TA | |||
| 2nd round | 3E12F | ATA TTA TCG CTG CCG TTT TTG CT | ||
| 3E12R | CTA AAC AAG TTA TTG TTA AAT TCG GAG | |||
| pfs47 | Pfs47_SNP707_F | GAA GAA ACT ATT GTA GAA TCT GGA AA | [15,31] | |
| Pfs47SNP725_R | AAG GCA TTT TTA TAA CCA CAT TAT TA | |||
| pvcsp | 1st round | VCS-OF | ATG TAG ATC TGT CCA AGG CCA TAA A | [24,32] |
| VCS-OR | TAA TTG AAT AAT GCT AGG ACT AAC AAT ATG | |||
| 2nd round | VCS-NF | GCA GAA CCA AAA AAT CCA CGT GAA AAT AAG | ||
| VCS-NR | CCA ACG GTA GCT CTA ACT TTA TCT AGG TAT | [24,33] | ||
| pvmsp1 F2 block | 1st round | VMI-O2F | GAT GGA AAG CAA CCG AAG AAG GGA AT | [24,32] |
| VMI-O2R | AGC TTG TAC TTT CCA TAG TGG TCC AG | |||
| 2nd round | VMI-N2F | AAA ATC GAG AGC ATG ATC GCC ACT GAG AAG | ||
| VMI-O2R | AGC TTG TAC TTT CCA TAG TGG TCC AG | |||
| pvmsp3α | 1st round | Pvmsp3a P1 | CAG CAG ACA CCA TTT AAG G | [3,12] |
| Pvmsp3a P2 | CCG TTT GTT GAT TAG TTG C | |||
| 2nd round | Pvmsp3a N1 | GAC CAG TGT GAT ACC ATT AAC C | ||
| pvmsp3β | 1st round | Pvmsp3b P1 | GTA TTC TTC GCA ACA CTC | |
| Pvmsp3b P2 | CTT CTG ATG TTA TTT CCA G | |||
| 2nd round | Pvmsp3b N1 | CGA GGG GCG AAA TTG TAA ACC | ||
| Pvmsp3b N2 | GCT GCT TCT TTT GCA AAG G | |||
| pvs47 | 1st round | CM 000453 Fwd | CAC ACC ACC GCA AAC AGG | [16] |
| CM 000453 Rev | GTG CAC ATT CCG CGG TTG | |||
| 2nd round | nst 1440 Fwd | GCG GTC CAC CCT AAC TGT AA | This study | |
| nst 1440 Rev | TGC TGC AAA CCA CAC ATG T | [34] | ||
| Sequencing primers | pPvs47-F | ATA TTT CCA ACG AAG CAT TTA TGC | ||
| pPvs47-R | TTT TCC ATT ATG CTC ACA AAC GC | |||
| Pvs47F2 | GAA GAA AGG GGA GGA CCA AG | [16] | ||
| pvs48/45 | 1st round | Pvs4845F | GGA ATA ATT TCG ACC ACT C | [35] |
| 887 | TCA GAA GTA CAA CAG GAG GAG CAC | [36] | ||
| 2nd round | 866 | ATG TTG AAG CGC CAG CTC GCC AA | ||
| 887 | TCA GAA GTA CAA CAG GAG GAG CAC | |||
| pvs48/45 | Sequencing primers | pPvs4845F | ATG GCC AAA GGA GAG GTC AAG TAC | [34] |
| pPvs4845R | TCG GCA GAT GCA AGT GAA GGA AGT C | |||
| pPvs48/45-6C-F | TCG GCA GAT GCA AGT GAA GGA AGT C | |||
| Pvs4845FF | TGT AAA ATC TGC GGA CGT GA | [16] | ||
| Pvs4845RR | CGGGTGCTTTAAAAATGGAA | |||
| 1001F | GAATGAGTTGCCCTGGGGAA | This study | ||
| 1025F | TGCCCGAGTGCTTCTTTCAA | |||
| 249R | TCCTGGGATCTTCTTCGGGA | |||
| 494R | AAGGCCACTCTTCCCTTCAC |
| Sample | Origin | pfcrt | pfmdr1 | pfhrp2 | pfhrp3 | pfama1 | pfs47 |
|---|---|---|---|---|---|---|---|
| M1 | Unknown | 72CVMNK76 | NFSND | Present | Present | New haplotype (Acc. No. PV567569) | 707C, 725C |
| Sample | Origin | pvcsp | pvmsp3a-N1 | pvmsp3a-N2 | pvmsp3b-N1 | pvmsp3b-N2 | pvmsp1(F2) | pvs47 | pvs48/45 |
|---|---|---|---|---|---|---|---|---|---|
| M2 | Afghanistan | VK247 type. New haplotype (Acc. No. PV567574) | - | PD * | New haplotype (Acc. No. PV590042) | PD | - | F22, F24, K27, D31, S57, S62, L82, D156, V230, M233, E240, I262, I273, A373 | E35, Y196, H211, K250, D335, E353, A376, I380, K390, K418 |
| M3 | Venezuela | - | PD | PD | New haplotype (Acc. No. PV590042) | PD | PD | 22L, F24; K27, D31, S57, S62, L82, D156, V230, M233, E240, I262, I273, A373 | E35, Y196, 211N, K250, D335, E353, A376, I380, K390, K418 |
| M4 | Unknown | VK210 type. New haplotype (Acc. No. PV567571) | New haplotype (Acc. No. PV590041) | PD | - | - | - | F22, F24; K27, D31, S57, S62, L82, D156, V230, M233, E240, I262, I273, A373 | - |
| M5 | Unknown | VK210 type. New haplotype (Acc. No. PV567570) | PD | PD | New haplotype (Acc. No. PV590042) | PD | PD | 22L, F24; K27, D31, S57, S62, L82, D156, V230, M233, E240, I262, I273, A373 | E35, Y196, 211N, K250, D335, E353, A376, I380, K390, K418 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godoy, A.; Euceda, K.; Pinto, A.; Valdivia, H.O.; Chaver, L.; Ardon, G.; Fontecha, G. Molecular Characterization of Plasmodium Species to Strengthen Malaria Surveillance in Migrant Populations in Honduras. Trop. Med. Infect. Dis. 2025, 10, 292. https://doi.org/10.3390/tropicalmed10100292
Godoy A, Euceda K, Pinto A, Valdivia HO, Chaver L, Ardon G, Fontecha G. Molecular Characterization of Plasmodium Species to Strengthen Malaria Surveillance in Migrant Populations in Honduras. Tropical Medicine and Infectious Disease. 2025; 10(10):292. https://doi.org/10.3390/tropicalmed10100292
Chicago/Turabian StyleGodoy, Ashley, Kevin Euceda, Alejandra Pinto, Hugo O. Valdivia, Lesly Chaver, Gloria Ardon, and Gustavo Fontecha. 2025. "Molecular Characterization of Plasmodium Species to Strengthen Malaria Surveillance in Migrant Populations in Honduras" Tropical Medicine and Infectious Disease 10, no. 10: 292. https://doi.org/10.3390/tropicalmed10100292
APA StyleGodoy, A., Euceda, K., Pinto, A., Valdivia, H. O., Chaver, L., Ardon, G., & Fontecha, G. (2025). Molecular Characterization of Plasmodium Species to Strengthen Malaria Surveillance in Migrant Populations in Honduras. Tropical Medicine and Infectious Disease, 10(10), 292. https://doi.org/10.3390/tropicalmed10100292








