Abstract
To better understand differential sensitivities among fish species to the piscicidal compound Antimycin-A (ANT-A), we hypothesized that variations in amino acids at the ANT-A binding site may reflect toxicity differences. Protein sequences for six motifs comprising the ANT-A binding site were obtained and compared for invasive carp species (N = 515) and seven non-target species (N = 277); a consensus was delineated from each species. The carp species, Common Carp (Cyprinus carpio), Silver Carp (Hypophthalmichthys molitrix), Bighead Carp (Hypophthalmichthys nobilis), Grass Carp (Ctenopharyngodon idella), and Black Carp (Mylopharyngodon piceus), showed the same amino acids at the site; thus, it was termed the carp consensus motif sequence (CCM). Channel Catfish (Ictalurus punctatus) showed the most amino acid polymorphisms, with three motifs 96–100% different from CCM. Within a species, Bluegill (Lepomis macrochirus) and Fathead Minnow (Pimephales promelas) variation per motif was most dissimilar (46.7% and 21.6%, respectively). Organismal mortality data from the literature indicated Yellow Perch (Perca flavescens), Walleye (Sander vitreus), and American Gizzard Shad (Dorosoma cepedianum) to be most sensitive to the piscicide, Catfish least sensitive, and all others intermediate. The protein sequence variations of the binding site appeared to be in accord with organismal sensitivity categories when they differed from the CCM; the motifs in Gizzard Shad and Walleye were the same as in CCM. The physical/chemical nature of ANT-A is important to consider in organismal response comparisons. This cellular approach of studying ANT-A binding at its target enzyme is a non-destructive way to predict piscicidal efficacy of ANT-A against fishes of interest, informs management decisions in control efforts for invasives, and can be used to forecast effects on sympatric species.
Keywords:
Antimycin-A; cytochrome b; invasive carp; amino acid polymorphism; mode of action; mitochondria Key Contribution:
Management may estimate levels of risk and tailor ANT-A applications in specific ecosystems where invasive carp occur with sympatric fish. With other invasive fish issues, ascertaining the amino acids per fish species at the six target sequences vital for ANT-A binding can inform an application approach.
1. Introduction
Reproductive capability, population densities, feeding habits, climate tolerances, mobility, and longevity are life history traits of invasive carp that underscore the ecological and economic concerns on behalf of the U.S. and Canada, especially when populations become established [1,2,3,4,5]. Of the carps native to Asia that have been introduced to the United States [4], four species of particular interest are Grass Carp (Ctenopharyngodon idella), Black Carp (Mylopharyngodon piceus), Bighead Carp (Hypophthalmichthys nobilis), and Silver Carp (H. molitrix). Grass Carp were initially imported into the U.S. as a means of biological control for nuisance aquatic vegetation and have established reproductive populations in the Lake Erie and Mississippi River basins [6,7]. Both diploids and triploids are widespread, considering the varying state laws surrounding permissions on commercially produced invasive carp [8] (Figure 1). Silver Carp and Bighead Carp feed at the base of the food chain [9,10]. They were introduced in the 1990s to reduce phytoplankton in sewage treatment ponds and enhance water quality in aquaculture ponds [3,11]. The first Black Carp from the wild was found on 26 March 2003 [12]. These benthic-feeding invertivores had been imported to control aquaculture pond snails, and since then, both fertile and functionally sterile individuals are established in parts of the Mississippi River Basin [13,14] (Figure 1b). They are feeding almost exclusively on mollusks and native freshwater mussels already in decline and may intensify that decline [5]. The latter three species are listed as injurious wildlife by the U.S. Fish and Wildlife Service under the Lacey Act (50 CFR §16) [15] (https://www.fws.gov/node/266035, accessed on 2 May 2023). The four invasive carp species are widespread nationally, particularly in the Mississippi Alluvial Basin.
Invasive carp eradication methods include physical removal, toxic baits, genetic technologies, and non-specific toxicant applications. The only successful integrated pest management strategy reported for an aquatic invasive species is the targeted toxicant 3-Trifluoromethyl-4-nitrophenol (TFM) used in controlling sea lamprey (Petromyzon marinus) in the Great Lakes [2,16]. A similar species-specific toxicant approach may be used to manage carp populations, perhaps in accordance with bio-acoustic tactics [17], feeding selective toxic baits [2,18,19], or waterway structure modifications [20]. Rotenone and Fintrol™ formulations (containing Antimycin-A; ANT-A) were two products registered by the United States Environmental Protection Agency (EPA) as piscicides for controlling bony fish [2,19], and in August 2017 the EPA registration expired for Fintrol’s use as a fish toxicant (https://ordspub.epa.gov/ords/pesticides/f?p=113:1::::1::) [21]. ANT-A is a mitochondrial inhibitor that interacts at a specific site in the series of proteins that make up the electron transport complex cytochrome bc1 or the ubiquinol:cytochrome c oxidoreductase membrane protein. With the binding of ANT-A, the enzyme is inhibited in its catalysis of electron transfer from ubiquinol to cytochrome c [22]. In that study of cytochrome bc1 structures of cow and chicken, the movement of an iron-sulphur protein was important in the electron transfer, with slight structural differences (e.g., transmembrane helix subunits), yet the binding site remained the same [22]. The amino acid sequence of cytochrome b among human to parasite species has been delineated in a study of mitochondrial respiratory chain inhibitors with the zebrafish model for mitochondria-linked disorders [23]. Cytochrome b is the only protein product of the mitochondrial genome that is a fully functional monomer and is used for developing markers for genetic stock identification, as a primary population genetic indicator in many fish species and for phylogenetic and taxonomic purposes, making it a pragmatic gene to study for a project such as this due to the amount of data already in the literature [24,25].
Thus, differences in the amino acid sequence of cytochrome b subunit Qi of complex III are functionally relevant for delineating differences among fish species at the target site of ANT-A binding. Using liver mitochondria of channel catfish (Ictalurus punctatus) and rainbow trout (Salmo gairdneri), a difference between species in the percent of inhibition by Ant-A concentrations was noted, with trout more affected [26]. Based on the literature and protein chemistry, the hypothesis in this study was that the amino acids of the binding site for ANT-A may be different among, but similar within, fish species. In support of implementing such goals of the Invasive Carp Action Plan as conducting research to provide accurate and scientifically valid information necessary for the effective management and control of invasive carps [4], the intent of this particular study was to identify the amino acids comprising the cytochrome bc1 binding site for ANT-A among invasive carp and some sympatric non-target species and to compile some published works regarding whole-fish lethality induced by various ANT-A exposure conditions (Table 1).

Table 1.
Published a differential lethality effects of Antimycin-A on selected fish species investigated in this study.
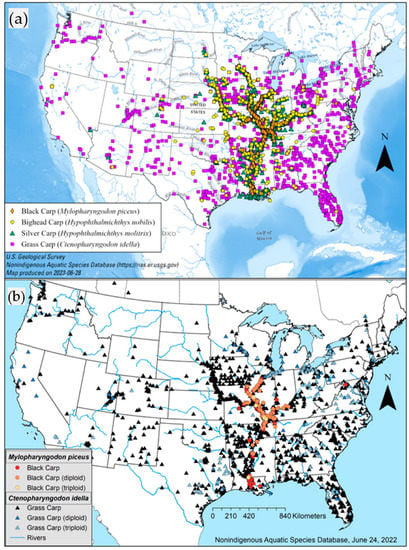
Figure 1.
(a) Map of historical invasive carp distributions by species in the U.S. [27]. (b) Map of historical captures of diploid and triploid Black Carp and Grass Carp in the U.S. [27].
Historically, research has been performed at the level of the organism for obtaining deleterious effects from ANT-A exposure, typically by lethality measures. Studies identified trends in sensitivities, with ANT-A being more toxic to scaled fishes [28,35]. Three categories were revealed: extremely sensitive (trout, perch, herring, and white suckers); intermediately sensitive fish (northern pike, sunfish, bigmouth buffalo, stickleback, minnows, and carp); and fish exhibiting little sensitivity (freshwater catfish) [26]. Mined from some of the literature, the ranges were noted in ANT-A concentrations estimated for organismal lethality effect (Table 1). These variable results reported are supported by reasons directly related to the physical/chemical properties of the ANT-A compound itself. Parameters that influence ANT-A stability include temperature, water pH, alkalinity, ultraviolet light, dissolved iron, formulations of antimycin used (which include diluents with variable surfactants and other compounds such as acetone, nonoxynol-9 detergent, diethyl phthalate, and soy lipids) holding tank systems for exposures, presence of debris such as organics, times applied in the exposures (e.g., longer exposure times require lower concentrations), exposure routes (degradation occurs after ingestion; carrier differences have been noted via ethanol or corn oil), and body mass not being considered per dose [18,33,36,37]. Moreover, antimycin is composed of four major isomers and at least four minor compounds; thus, its manufacture, which includes fermentation and refinements, influences its isomeric compositional mix and quality [37,38]. Slight structural features, such as side chains on antimycin isomer 1, can influence binding [39]; however, this influence is measurable only at the atomic level and not relevant for considerations of ANT-A binding at a broader scale.
Gaining a greater understanding of species differences and potential sensitivities at the molecular level should shed light on reported variabilities in organismal mortalities and can point to further investigations in a more targeted methodology (Table 1). As with TNF used in an integrated pest management approach, ANT-A used in conjunction with other control efforts such as herding [40] and acoustic deterrents or attractants [17] may be considered for invasive carp management [41]. If ANT-A is to be considered for EPA re-registration as a water additive or for use in an oral delivery formulation [2], data at the cell and molecular level are relevant. Our hypothesis was that differences in organismal sensitivities to ANT-A among species—tested under consistent environmental conditions without interfering factors described—are related to the primary protein structure of the ANT-A binding site and that these amino acid sequences and subsequent protein conformational differences or similarities are relevant for ANT-A binding.
Like rotenone, ANT-A inhibits respiration in sensitive organisms, with both compounds believed to be rapidly absorbed from the water across the gills as the mode for entering the bloodstream [35]. Rotenone is a mitochondrial respiratory complex I inhibitor, and ANT-A is a complex III inhibitor [23]. The toxicity of ANT-A to fish is species-dependent and varies widely (Table 1) [29,35], likely because of subtle species differences in the protein sequences to which it binds [37]. The attribute of differential responses at different application rates and mechanisms allows ANT-A to be used as a selective toxin [37]. Antimycin-A interacts at the Qi subunit of cytochrome b located in the third mitochondrial protein complex in the series that comprises the electron transport chain in the inner mitochondrial membrane (Figure 2). The ANT-A molecule binds tightly to subunit Qi and prevents ubiquinol (also called coenzyme Q) from binding to cytochrome b, blocking the shuttle of electrons and halting ATP production.
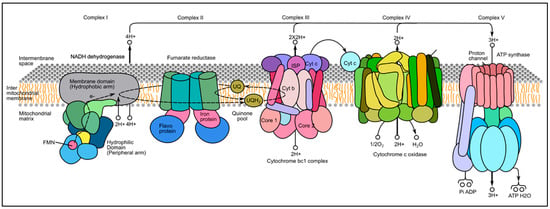
Figure 2.
The series of proteins comprising the electron transport chain in the inner mitochondrial matrix. The target binding site for Antimycin-A is the Qi subunit within cytochrome b (complex III). Image by M. Martinez as modified from [42].
This induces mitochondrial dysfunction and causes the bound oxygen to convert to superoxide that builds up at an accelerated rate, overwhelming the cell. Both the electron shuttle inhibition and the superoxide generation cause subsequent cell death [37,43]. Cytochrome b within complex III (cytochrome bc1 complex) is within the inner mitochondrial matrix of most cell types [23,43] but not in non-nucleated mammalian red blood cells.
2. Materials and Methods
Amino acid sequence records for the Qi subunit of cytochrome b were obtained from the protein database of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/protein/?term=cytochrome+B+subunit+qi) (accessed on 21 May 2019 to 17 June 2019) [44], yielding multiple accession identifiers per species for Common Carp (Cyprinus carpio), Silver Carp (Hypophthalmichthys molitrix), Bighead Carp (Hypophthalmichthys nobilis), Grass Carp (Ctenopharyngodon idella), Black Carp (Mylopharyngodon piceus), Fathead Minnow (Pimephales promelas), Bluegill (Lepomis macrochirus), Channel Catfish (Ictalurus punctatus), Yellow Perch (Perca flavescens), Walleye (Sander vitreus), Nile Tilapia (Oreochromis niloticus), and American Gizzard Shad (Dorosoma cepedianum) (Supplementary Table S1). The compiled accession identification amino acid sequences were imported into Genomics Workbench 21 Software (QIAGEN; Redwood City, CA) aligned, and analyzed by the six sequence motifs that conformationally fold to form the 3-D protein structure in complex III, where ANT-A (CAS 1397-94-0) binds [23] (Figure 3).
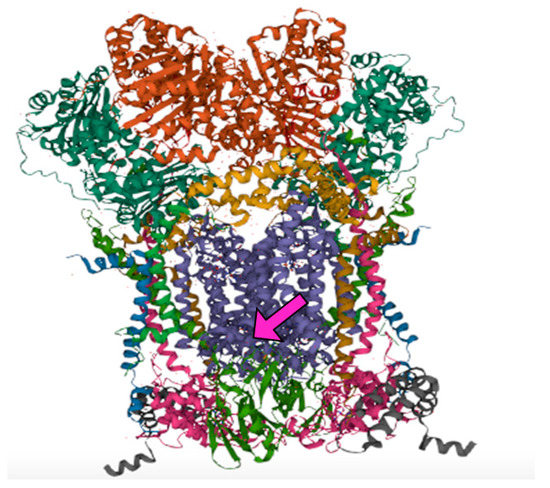
Figure 3.
Secondary protein structure of antimycin A1 in complex with cytochrome bc1 as created with Visual Molecular Dynamics molecular modeling and visualization software v. 1.9.3 (Urbana, IL). The Antimycin-A interaction (arrow) is at the ligands in the lower portion of the bc1 complex.
Investigating the potential amino acid polymorphisms for the six motifs, both within and among species, illuminated similarities and variations. Some of the NCBI accession identifiers for individuals did not include all six motifs (the full complement of amino acids for all motifs); thus, the formulae defined below considered only accessions where the full complement of amino acids was presented. Within a species, a potential consensus of amino acids for all six motifs was investigated and derived and, if found, was termed a species-specific consensus motif sequence (SSCM). Because each of the carp species displayed the same amino acid sequences in each of the six motifs comprising the ANT-A binding site (Table 2), these motifs were collectively termed carp consensus motif sequence (CCM). This CCM was compared with the non-carp motifs, and the amino acids not matching CCM were noted (Table 2).

Table 2.
Details of amino acid sequence accessions for the Qi subunit of cytochrome b obtained from the National Center for Biotechnology Information (NCBI) protein database for selected fish species. The motifs column indicates the species-specific consensus motif sequence (SSCM) of each species. Amino acid abbreviations in bold, red, and italics are those that differed from the carp consensus motif sequence (CCM), where each of the six motifs for the carp was the same.
3. Results
Each species displayed an SSCM, which was the most frequently occurring amino acid sequence in the six motifs (Table 2). Some variation did occur simply due to individual accessions having a single amino acid variation, which can happen due to anatomical variation, laboratory methodology issues, a single undefined amino acid (denoted in NCBI as X; 16 amino acids in Common Carp, 1 Bighead Carp, and 1 Silver Carp), or possible human errors in editing original accessions. In this manuscript, because all carp species showed a consensus motif sequence, it is being defined as the carp consensus motif sequence, or CCM, as previously stated in the methods and used in the formulae. Intraspecies variation indicated whether protein sequences varied at the ANT-A binding site within a species, per the delineated motifs (Formula (1); Figure 4; Table 2). These data indicate the likelihood that all individuals of a species presented the same amino acid per motif. Among the 12 species, four of the six motifs showed within-species variation (Figure 4). Bluegill and Fathead minnow showed the highest levels of intraspecies variation, with 46.7% and 21.6% variation within a single but different motif, respectively (Figure 4).
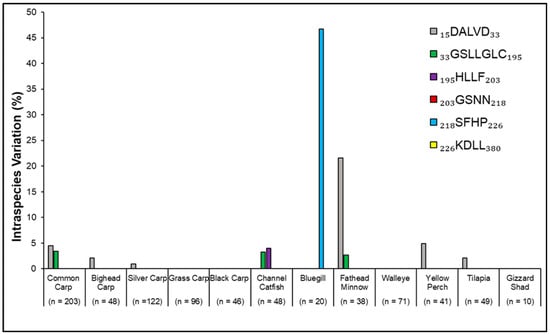
Figure 4.
Within-species comparisons of the six amino acid sequences (motifs) comprising the Antimycin-A binding target site in the mitochondrial cytochrome b subunit Qi of complex III showed the percent intraspecies variation per motif. Motifs are denoted by their abbreviations, with sequences having been obtained from a National Center for Biotechnology Information (NCBI) Protein Blast search. Sequence location within the collective motif is delineated by the two numerical values on either side of the abbreviated name in the legend. Sample size (n) denotes the total number of accessions available and analyzed per species. All motifs are shown in the key, even those that did not exhibit intraspecies variation.
Data from the NCBI accession identifiers were used in formulae to quantify similarities and differences in amino acid sequences for the Qi subunit of cytochrome b. The denominator term “complete motifs” indicates the number of sequences among the accession identifiers that included the full complement of amino acids. Delineation of the percent of variation within a species (intraspecies variation) per motif for the twelve chosen fish species (Figure 4) was calculated as Formula (1):
Per motif among the twelve fish species, the percent of variation among species (interspecies variation) was calculated. Thus, this value reflected how much that motif varied in relation to the complete motifs delineated for all individuals among the twelve species (Figure 5) performed as Formula (2):
Which of the six motifs among all species displayed the most variation as compared to the CCM motif (Figure 6) was calculated as Formula (3):
Which of the six motifs among non-carp species displayed variation as compared to the CCM motif (Figure 7) was determined by Formula (4):
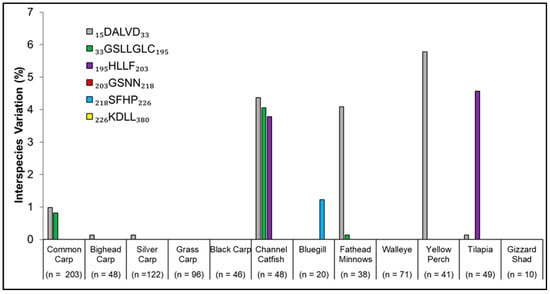
Figure 5.
Among-species comparison of individual motifs comprising the Antimycin-A binding target site in the mitochondrial cytochrome b subunit Qi of complex III. Percent of variation among species was obtained using Formula (1). Amino acids are denoted in the legend by their abbreviations, with sequences having been obtained from a National Center for Biotechnology Information (NCBI) Protein Blast search. The motif location within the collective protein sequence is delineated by the numerical values on either side of the motif abbreviation. Sample size (n) denotes the total number of individuals available to analyze per species. All motifs are shown in the key, even those that did not exhibit interspecies variation.
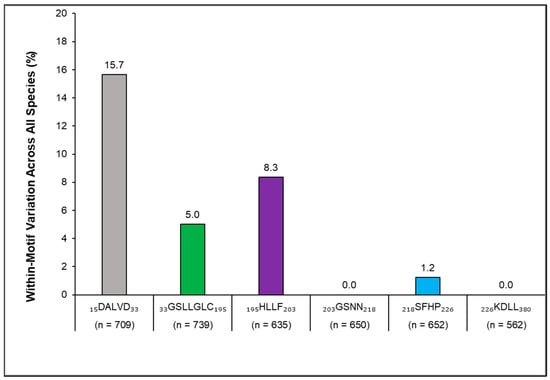
Figure 6.
Per-motif variation from the carp consensus motif sequence (CCM) was generated for all sympatric species (Fathead Minnow (Pimephales promelas), Bluegill (Lepomis macrochirus), Channel Catfish (Ictalurus punctatus), Yellow Perch (Perca flavescens), Walleye (Sander vitreus), Nile Tilapia (Oreochromis niloticus), and American Gizzard Shad (Dorosoma cepedianum)). The percent variation was highest for the first of the six motifs that comprise the Antimycin-A-binding target site in the mitochondrial cytochrome b subunit Qi of complex III. Sequence location within the collective protein sequence is delineated by the numerical values on either side of the motif abbreviation. Sample size (n) denotes the total number of amino acids analyzed per motif.
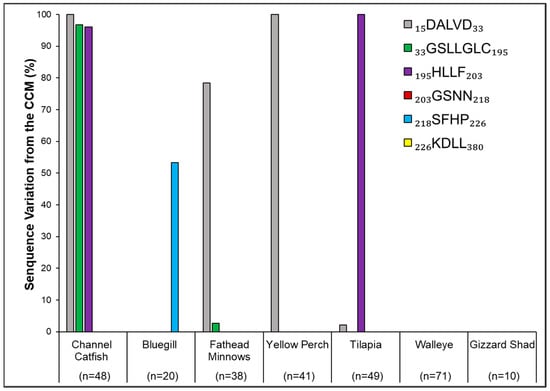
Figure 7.
Per-motif amino acid sequence variations within non-carp fish species compared to the carp consensus motif sequence (CCM) in relation to the total number of individuals within that species. Sample size (n) denotes the total number of individuals available and analyzed per species. All motifs are shown in the key, even those that did not exhibit sequence variation.
Interspecies variation indicated the number of amino acid polymorphisms per motif per species that differed from the CCM as compared to the complete number of motifs across all individuals over all species (Formula (2)). All individuals from Grass Carp, Black Carp, Walleye, and American Gizzard Shad showed no variability in any of the motifs that comprise the ANT-A binding site (Formula (2); Figure 5). Of all species, Channel Catfish showed the most interspecific variation; three motifs showed variability compared to CCM. Among all species, Yellow Perch displayed the most variation in a single motif at 5.8% in the first motif, which was also the motif displaying dissimilarity in 53.8% of the times (7 of 13) that variation across all species was noted (Figure 5).
Focusing on which of the motifs varied the most among the species researched, the percent of amino acid variation per motif was calculated as compared with CCM (Formula (3); Figure 6). The first motif along the protein’s progression of six amino acid sequence motifs in the mitochondrial cytochrome b subunit Qi of complex III making up the ANT-A binding site displayed the highest percent variation (15.7%), followed by the third motif (8.4%), the second (5.0%), and then the fifth motif (1.2%). The fourth and sixth motifs exhibited no variation in any of the individuals among the species analyzed (Figure 6).
Within non-carp species, variations per motif in relation to the CCM (Formula (4); Figure 7; Table 2) illustrated that Walleye and Gizzard Shad showed complete similarity with CCM. The other five species showed considerable variation, with Channel Catfish showing the most and with variability in the first 3 of the 6 motifs being dissimilar from CCM at 98% of the individual identifiers (Figure 7). See Table A1 for the locations of the amino acids in non-carp species that differed from CCM. See Supplemental Table S2 for a summary of specifics per species.
4. Discussion
Effective tools can help resource managers control invasive and nuisance fishes, often including overharvest, toxicants, and acoustic deterrents [45,46]. Although drawbacks of ANT-A use include adverse chemical residues remaining in fish and possibly the need for chemical detoxification of waterways after application and keeping the public away [45], its effectiveness, utility, and attributes suggest that the compound may warrant further detailed study in light of reregistration efforts. Managerial benefits of ANT-A use include that concentrations for cyprinid mortality are lower than that for rotenone [30,38], doses required to eliminate fish do not harm higher vertebrates or invertebrates [47], ANT-A is susceptible to decomposition and degrades rapidly [35,37], and fishes can voluntarily feed on impregnated bait [48].
In this study, both Walleye and Gizzard Shad showed species-specific consensus motifs identical to the CCM, indicating a consensus among the ANT-A binding site and thus suggesting similar susceptibilities to ANT-A as targeted invasive carp. In such cases, additional considerations for fish management strategies would be appropriate when desirable species occur in the same aquatic ecosystem. Further molecular investigations may reveal species differences, but if this deduced lethality response is predicted, some alternatives could be to capture fish at pre-treatment and or stocking/restocking efforts post-treatment.
Although models for ANT-A binding within cytochrome b have shown the importance of the formyl salicylic acid and dilactone portions of the molecule [49], investigations into amino acid polymorphism effects on mitochondrial function and resultant organismal toxicity have not yet been performed among fish species. Zebrafish (Danio rerio) embryos were used to test mitochondrial respiratory chain inhibitors, including ANT-A, so as to address human mitochondrial-linked disorders and antiparasitic drugs; complex III inhibitors induced mortality more quickly than the complex I inhibitors (e.g., rotenone), which induced more abnormalities in survivors [23]. The fact that abnormality inductions did not occur in early-life stages [23] might also be considered an asset of ANT-A use over rotenone. To provide context about the amino acid sequence similarity of cytochrome b (complex III) of humans compared with mice, zebrafish, fungi, and a protozoan species, variability ranged from 79%, 74%, 56%, and 41%, respectively [23]. The similarity among teleosts shown by our study would thus be expected to be much higher, and the variabilities more subtle than the between-taxon comparisons of Complex III inhibitors. Measuring respiratory effects directly on mitochondrial function in blood cells of target carp and sympatric fishes would be a direct measure of the effect that would occur prior to whole-body respiratory effects/lethality, with respect to consistent and repeatable experimental procedures with ANT-A. The mitochondrial electron transport complex of proteins is very similar across all species that utilize oxygen [37] and in the gene coding for cytochrome b [24,25]; thus, the actual amino acids comprising the ANT-A binding location must be evolutionarily conserved.
The challenging chemistry of the ANT-A compound has been a deterrent for in vitro studies, with many more being conducted at the organismal level (Table 1). As described earlier, several parameters influence ANT-A stability, including it decays exponentially over time with rates as a function of pH, temperature, and water hardness [37]. Most, if not all, field studies have not tracked and recorded such variables. According to these studies based on whole fish responses, the lethality of ANT-A varies from <1.0 µg/L for most trout and char (Salmonidae) to 25–200 µg/L for most freshwater catfish (Ictaluridae) [35]. Most minnows (Cyprinidae) and sunfish (Centrarchidae) were estimated to suffer mortality at 5.0–10 µg/L [29,35] (Table 1). ANT-A was estimated to be 1000 times more toxic to sensitive species than to resistant ones, in order of decreasing sensitivity: Rainbow Trout, Bluegill, Green Sunfish, Goldfish, Channel Catfish, and Black Bullhead [36]. In that study, the LC50s ranged from 50 ppb to 50 ppt for black bullhead and rainbow trout, respectively, in neutral pH waters. Because ANT-A degrades more rapidly in alkaline pH [36], future in vitro studies performed at neutrality could help in a variety of aquatic ecosystems with species of interest.
Looking at the variation among species and the six motifs comprising the binding site, the most variation was in the non-carp, whereby 54% of the NCBI database accessions were different (277 out of 515) (Figure 5). Because the first motif along the protein’s progression of six amino acid sequence motifs displayed the highest percent variation (Figure 6), future amino acid sequence searches might focus heavily on this region, as well as the other three that showed amino acid differences from the CCM. All carp, along with Walleye and Gizzard Shad, showed a low percent (Common, Bighead, and Silver Carp) to zero percent interspecies variation (Grass Carp, Black Carp, Walleye, Gizzard Shad) (Figure 5 and Figure 7). Catfish are known to be some of the least susceptible fish to ANT-A, especially in comparison to carp sensitivity [33]. Compared to the invasive carp CCM, the Channel Catfish motifs showed the highest level of amino acid variations and within the most motifs (Figure 7). This universal consensus of sequences derives from the criticality of the motifs in their function for cellular respiration. Because of this similarity shown in these seven species, upon in vitro testing, mitochondrial inhibitory responses to ANT-A concentrations may be anticipated to be consistent among individuals.
In examining the characteristic changes in amino acids that varied from the CCM within each species, the Fathead Minnow, Bluegill, Yellow Perch, and Channel Catfish variant displayed a change in polarity, whereas Nile tilapia did not (Table 3). Polar amino acids are hydrophilic in nature, and non-polar amino acids are hydrophobic; these characteristics, in addition to other secondary structure characteristics [50], contribute to protein structure and function and a loss or gain of polarity and/or charge in a protein’s structure will therefore influence the ability of ANT-A to bind. Channel Catfish and Yellow Perch are described as displaying the least and greatest susceptibility to ANT-A, respectively; both species’ variants showed polarity changes in the amino acids that differed from the CCM, with the highest number of different amino acids in catfish. A change to the primary structure of a protein can cause a conformational change in the secondary, tertiary, and quaternary structure of a protein and, therefore, affect protein structure and function.

Table 3.
Characteristics of the amino acids in the non-carp species that varied from the CCM.
Three-dimensional structures, which are in turn determined by their genetically encoded amino acid sequences, characterize the proteins structurally and functionally [50,51]. Amino acid variations that were deduced from nucleotides from PCR products of cytochrome b of Korean salmonids were used for distinguishing systematic relationships, as most of the nucleotide substitutions did not alter the amino acid sequence of the protein [24]. In this study, modeling potential tertiary protein structures for cytochrome bc by using databases and tools for protein predictions [50,51] would help in making connections with ANT-A’s ability to bind and, therefore, its effectiveness. Because the protein prediction tools typically do not include fish species outside laboratory models, a PCR-based approach to delineating nucleotides of cytochrome bc and then deducing amino acid sequences [25] for species of interest is a feasible approach for contributing data on the amino acid sequences for their binding motifs. The molecular characteristics of the amino acid variants from the CCM (Table 3) provide a weight of evidence to the hypothesis that ANT-A’s effectiveness is predictable by protein structure. Because Channel Catfish, Yellow Perch, Nile Tilapia, Bluegill, and Fathead Minnows have high percent variation from the CCM, this indicates that these species likely have different susceptibilities to ANT-A than carp, whereas Walleye and Gizzard Shad are unlikely to have differing sensitivities. The amino acid positions of variation in each of the non-carp species (Table 3) and the classification of both the CCM and the variable amino acids within non-carp species that are likely to have differing sensitivities (Table 3) will inform further investigations of ANT-A exposure results for each of these species. Thus, data generated at the cell and molecular level of this mitochondrial respiratory chain inhibitor can provide an additional basis by which to assess the efficacy of the ANT-A or its formulation, as well as organismal effects on sympatric species. This approach of determining amino acid positions vital to the binding of ANT-A can be considered in other fisheries management issues using this piscicide, such as for potential control of non-native smallmouth bass (Micropterus dolomieu) proliferation below the Glen Canyon Dam in the Glen Canyon National Recreation Area as a threat to humpback chubs (Gila cypha), for example. Regarding invasive carp and sympatric species, when more variation occurs in the CCM than in the non-targets, management may predict levels of risk or may tailor ANT-A applications in specific aquatic ecosystems where target invasive carp occur.
5. Conclusions
To better understand differential toxicities among fish species, we mined protein sequence data from the National Center for Biotechnology Information and compared the amino acid sequence data for the six motifs that comprise the ANT-A target binding site among and within invasive carp species and some non-targets. The carp species (Common Carp, Silver Carp, Bighead Carp, Grass Carp, and Black Carp) showed the same amino acids at the site; thus, it was termed the carp consensus motif sequence (CCM). Channel Catfish showed the most amino acid polymorphisms. Although the protein sequence data variations for the binding site appeared to be in accord with the organismal sensitivity categories, with ANT-A being more toxic to scaled fishes, the challenging chemistry of the ANT-A compound can interfere with organismal studies. Thus, such a comparative approach is relevant for delineating potential differences in the toxicity of ANT-A to fishes of interest.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes8070381/s1; Table S1: Accession numbers for amino acid sequence records for the Qi subunit cytochrome b obtained (5/21/2019 to 6/17/2019) from the protein database of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/protein/?term=cytochrome+B+subunit+qi) from several fish species used in this study; Table S2: Intraspecies and Interspecies amino assay variation.
Author Contributions
Conceptualization, J.A.J.; methodology, B.A.B. and B.L.B.; validation, B.A.B., B.L.B. and J.A.J.; formal analysis, B.A.B. and B.L.B.; investigation, B.A.B. and B.L.B.; resources, J.A.J.; data curation, B.L.B.; writing—original draft preparation, B.A.B.; writing—review and editing, B.A.B., J.A.J. and B.L.B.; visualization, B.A.B., J.A.J. and B.L.B.; supervision, R.D.C.; project administration, J.A.J.; funding acquisition, R.D.C. and J.A.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by U.S. Geological Survey Ecosystems Mission Area’s Environmental Health Program and the Biological Threats and Invasive Species Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: https://www.ncbi.nlm.nih.gov/(accessed on 21 May 2019 to 17 June 2019).
Acknowledgments
The authors thank Wesley Daniel of the U.S. Geological Survey for producing current maps for Figure 1, Melinda Martinez (USGS) for Figure 2, Kelly Wood (USGS) and Lydia Duhon (USGS) for assisting with Table 1 and references, and Joel Putnam, formerly with the USGS, for reviewing an earlier draft of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Locations of amino acids in non-carp species that differed from the carp consensus motif (CCM) within the Antimycin-A binding site motifs are noted in bold. The CCM amino acid is provided.
Table A1.
Locations of amino acids in non-carp species that differed from the carp consensus motif (CCM) within the Antimycin-A binding site motifs are noted in bold. The CCM amino acid is provided.
| Fish | Amino Acid Position | ||||||
|---|---|---|---|---|---|---|---|
| Common Name | Scientific Name | 16 | 19 | 38 | 197 | 198 | 219 |
| Fathead Minnow | Pimephales promelas | Gly | Val | Gly | Leu | Leu | Ser |
| Bluegill | Lepomis macrochirus | Asp | Val | Gly | Leu | Leu | Leu |
| Yellow Perch | Perca flavescens | Asn | Val | Gly | Leu | Leu | Ser |
| Nile Tilapia | Oreochromis niloticus | Asp | Val | Gly | Leu | Ile | Ser |
| Channel Catfish | Ictalurus punctatus | Asn | Ile | Leu | Ala | Leu | Ser |
References
- Wittmann, M.E.; Jerde, C.L.; Howeth, J.G.; Maher, S.P.; Deines, A.M.; Jenkins, J.A.; Whitledge, G.W.; Burbank, S.R.; Chadderton, W.L.; Mahon, A.R. Grass carp in the Great Lakes region: Establishment potential, expert perceptions, and re-evaluation of experimental evidence of ecological impact. Can. J. Fish. Aquat. Sci. 2014, 71, 992–999. [Google Scholar] [CrossRef]
- Poole, J.R.; Sauey, B.W.; Amberg, J.J.; Bajer, P.G. Assessing the efficacy of corn-based bait containing antimycin-a to control common carp populations using laboratory and pond experiments. Biol. Invasions 2018, 20, 1809–1820. [Google Scholar] [CrossRef]
- Kolar, C.S.; Chapman, D.C.; Courtenay, W.R.; Housel, C.M.; Williams, J.D.; Jennings, D.P. Bigheaded Carps: A biological Synopsis and Environmental Risk Assessment; American Fisheries Society: Bethesda, MD, USA, 2007; 204p. [Google Scholar]
- Conover, G.; Simmonds, R.; Whalen, M. (Eds.) Management and Control Plan for Bighead, Black, Grass, and Silver Carps in the United States; Asian Carp Working Group, Aquatic Nuisance Species Task Force: Washington, DC, USA, 2007; 223p. [Google Scholar]
- Cudmore, B.; Jones, L.A.; Mandrak, N.E.; Dettmers, J.M.; Chapman, D.C.; Kolar, C.S.; Conover, G. Ecological Risk Assessment of Grass Carp (Ctenopharyngodon idella) for the Great Lakes Basin. Fisheries and Oceans Canada, Canadian Science Advisory Secretariat: Ottawa, ON, Canada, 2019; 115p. [Google Scholar]
- Conner, J.V.; Gallagher, R.P.; Chatry, M.F. Larval evidence for natural reproduction of the grass carp (Ctenopharngodon idella) in the lower Mississippi River. In Proceedings of the Fourth Annual Larval Fish Conference, University of Mississippi, Oxford, MS, USA, 27–28 February 1980; Fuiman, L.A., Ed.; FWS/OBS-80/43; Fish and Wildlife Service: Washington, DC, USA, 1980; pp. 1–19. Available online: http://pubs.er.usgs.gov/publication/fwsobs80_43 (accessed on 28 June 2023).
- Chapman, D.C.; Davis, J.J.; Jenkins, J.A.; Kocovsky, P.M.; Miner, J.G.; Farver, J.; Jackson, P.R. First evidence of grass carp recruitment in the Great Lakes Basin. J. Great Lakes Res. 2013, 39, 547–554. [Google Scholar] [CrossRef]
- Jenkins, J.A.; Chauvin, M.D.; Johnson, D.; Brown, B.L.; Bailey, J.; Kelly, A.M.; Kinter, B.T. Defensible standardized ploidy assessments for Grass Carp (Ctenopharyngodon idella, Cyprinidae) intercepted from the commercial supply chain. J. Great Lakes Res. 2019, 45, 371–383. [Google Scholar] [CrossRef]
- DeBoer, J.A.; Anderson, A.M.; Casper, A.F. Multi-trophic response to invasive silver carp (Hypophthalmichthys molitrix) in a large floodplain river. Freshw. Biol. 2018, 63, 597–611. [Google Scholar] [CrossRef]
- Zhang, H.; Rutherfor, E.S.; Mason, D.M.; Breck, J.T.; Wittmann, M.E.; Cooke, R.M.; Lodge, D.M.; Rothlisberger, J.D.; Zhu, X.; Johnson, T.B.; et al. Forecasting the impacts of silver and bighead carp on the Lake Erie food web. Trans. Am. Fish. Soc. 2016, 145, 136–162. [Google Scholar] [CrossRef]
- Kolar, C.S.; Courtenay, W.R.; Nico, L.G. Managing undesired and invading fishes. In Inland Fisheries Management in North America, 3rd ed.; Hubert, W.A., Quist, M.C., Eds.; American Fisheries Society: Bethesda, MD, USA, 2010; pp. 213–259. [Google Scholar]
- Chick, J.H.; Maher, R.J.; Burr, B.M.; Thomas, M.R. First black carp captured in the U.S. Science 2003, 300, 1875–1876. [Google Scholar] [CrossRef]
- Nico, L.G.; Jelks, H.L. The black carp in North America: An update. In Invasive Asian Carps in North America; Chapman, D.C., Hoff, M.F., Eds.; American Fisheries Society: Bethesda, MD, USA, 2011; Volume 74, pp. 89–104. [Google Scholar]
- Whitledge, G.W.; Kroboth, P.T.; Chapman, D.C.; Phelps, Q.E.; Sleeper, W.; Bailey, J.; Jenkins, J.A. Estabishment of invasive Black Carp (Mylopharyngodon piceus) in the Mississippi River Basin: Identifying sources and year classes contributing to recruitment. Biol. Invasions 2022, 24, 3885–3904. [Google Scholar] [CrossRef]
- Office of the Federal Register. Title 50—Wildlife and Fisheries.Chapter 1—United States Fish and Wildlife Service, Department of the Interior.Part 16—Injurious Wildlife; 50CFR16.13; U.S. Government Publishing Office: Washington, DC, USA, 1996; pp. 1–7.
- Hubert, T.D. Environmental fate and effects of the lampricide TFM: A review. J. Great Lakes Res. 2003, 29, 456–474. [Google Scholar] [CrossRef]
- Willis, D.J.; Hoyer, M.V.; Canfield, D.E.; Lindberg, W.J. Training grass carp to respond to sound for potential lake management uses. N. Am. J. Aquac. 2002, 22, 208–212. [Google Scholar] [CrossRef]
- Kroboth, P.T.; Chapman, D.C.; Steevens, J.A.; Byrd, C.G. Ingested toxicity of antimycin A to grass carp Ctenopharyngodon idella and black carp Mylopharyngodon piceus in two carriers. Manag. Biol. Invasions 2022, 13, 737–749. [Google Scholar] [CrossRef]
- Rach, J.J.; Boogaard, M.; Kolar, C. Toxicity of rotenone and antimycin to silver carp and bighead carp. N. Am. J. Fish. Manag. 2009, 29, 388–395. [Google Scholar] [CrossRef]
- Zielinski, D.P.; Sorensen, P.W. Numeric simulation demonstrates that the upstream movement of invasive bigheaded carp can be blocked at sets of Mississippi River locks-and-dams using a combination of optimized spillway gate operations, lock deterrents, and carp removal. Fishes 2021, 6, 10. [Google Scholar] [CrossRef]
- Department of Prevention, Pesticides and Toxic Substances. Reregistration Eligibility Decision for Antimycin A.; U.S. Environmental Protection: Washington, DC, USA, 2007; 34p.
- Zhang, Z.; Huang, L.; Shulmeister, V.M.; Chi, Y.; Kim, K.K.; Hung, L.W.; Crofts, A.R.; Berry, E.A.; Kim, S.H. Electron transfer by domain movement in cytochrome bc1. Nature 1998, 392, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Pinho, B.R.; Santos, M.M.; Fonseca-Silva, A.; Valentao, P.; Andrade, P.B.; Oliveira, J.M.A. How mitochondrial dysfunction affects zebrafish development and cardiovascular function: An in vivo model for testing mitochondria-targeted drugs. Br. J. Pharmacol. 2013, 169, 1072–1090. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lee, H.J.; Kim, W.J.; Lee, J.H.; Min, K.S. Mitochondrial cytochrome b sequence variation in Korean salmonids. J. Fish Biol. 2020, 56. [Google Scholar] [CrossRef]
- Khalifa, M.A.; Younes, M.I.; Ghazy, A. Cytochrome b shows signs of adaptive protein evolution in Gerbillus species from Egypt. J. Basic Appl. Zool. 2018, 79, 1. [Google Scholar] [CrossRef]
- Smith, M.J.; Simco, B.A.; Warren, C.O. Comparative effects of antimycin A on isolated mitochondria of channel catfish (Ictalurus punctatus) and rainbow trout (Salmo gairdneri). Comp. Biochem. Physiol. 1975, 52, 113–117. [Google Scholar] [CrossRef]
- U.S. Geological Survey. Nonindigenous Aquatic Species Database. Available online: https://nas.er.usgs.gov (accessed on 22 June 2022 and 28 June 2023).
- Walker, C.R.; Lennon, R.E.; Berger, B.L. Preliminary Observations on the Toxicity of Antimycin A to Fish and Other Aquatic Animals; Investigations in Fish Control; U.S. Department of the Interior, Bureau of Sport Fisheries and Wildlife: Washington, DC, USA, 1964.
- Berger, B.L.; Lennon, R.E.; Hogan, J.W. Ecotox: Laboratory Studies on Antimycin A as a Fish Toxicant; Investigation in Fish Control No.26, Fish Wildl. Serv., Bur. Sport Fish. Wildl.; USDI: Washington, DC, USA, 1969; 21p.
- Marking, L.L.; Bills, T.D. Sensitivity of four species of carp to selected fish toxicants. N. Am. Jounal Fish. Manag. 1981, 1, 51–54. [Google Scholar] [CrossRef]
- Gilderhus, P.A. Exposure times necessary for antimycin and rotenone to eliminate certain freshwater fish. J. Fish. Res. Board Can 1972, 29, 199–202. [Google Scholar] [CrossRef]
- Mayer, F.L.; Ellersieck, M.R. Manual of Acute Toxicity: Interpretation and Data Base for 410 Chemicals and 66 Species of Freshwater Animals; Technical Report PB86239878; U.S. Department of the Interior Fish and Wildlife Service: Columbia, MO, USA, 1986; USDI Fish and Wildlife Service, Publication No.160, Washington, DC; 588p. Available online: https://ntrl.ntis.gov/NTRL/dashboard/searchResults/titleDetail/PB86239878.xhtml# (accessed on 28 June 2023).
- Chapman, D.; Fairchild, J.; Carollo, B.; Deters, J.; Feltz, K.; Witte, C. An Examination of the Sensitivity of Bighead Carp and Silver Carp to Antimycin A and Rotenone; US Geological Survey: Columbia, MO, USA, 2003; pp. 1–22.
- Henderson, S. Preliminary studies on the tolerance of the White Amur, Ctenopharyngodon idella, to Rotenone and other commonly used pond treatment chemicals. In Proceedings of the Twenty-Seventh Annual Conference of the Southeastern Association of Game and Fish Commissioners, Hot Springs, AR, USA, 14–17 October 1973; United States Environmental Protection Agency: Washington, DC, USA, 1974; Volume 27, pp. 435–447. [Google Scholar]
- Finlayson, B.J.; Schnick, R.A.; Cailteux, R.L.; DeMong, L.; Horton, W.D.; McClay, W.; Thompson, C.W. Assessment of antimycin a use in fisheries and its potential for reregistration. Fisheries 2002, 27, 10–18. [Google Scholar] [CrossRef]
- Marking, L.L.; Dawson, V.K. The half-life of biological activity of antimycin determined by fish bioassay. Trans. Am. Fish. Soc. 1972, 101, 100–105. [Google Scholar] [CrossRef]
- Ott, K.C. Antimycin. A brief review of it’s chemistry, environmental fate, and toxicology. Biochem. Et Biophys. Acta 1994, 1185, 1–9. [Google Scholar]
- Clearwater, S.J.; Hickey, C.W.; Martin, M.L. Overview of Potential Piscicides and Molluscicides for Controlling Aquatic Pest Species in New Zealand; Department of Conservation: Wellington, New Zealand, 2008; p. 74.
- Kim, H.; Esser, L.; Hossain, M.B.; Xia, D.; Yu, C.; Rizo, J.; van der Helm, D.; Deisenhofer, J. Structure of antimycin A1, a specific electron transfer inhibitor of Ubiquinol−Cytochrome c Oxidoreductase. J. Am. Chem. Soc. 1999, 121, 4902–4903. [Google Scholar] [CrossRef]
- Ridgway, J.L.; Lawson, K.M.; Shier, S.A.; Calfee, R.D.; Chapman, D.C. An assessment of fish herding techniques: Management implications for mass removal and control of silver carp. N. Am. J. Fish. Manag. 2023, 43, 176–188. [Google Scholar] [CrossRef]
- Chapman, D.C. "Modified Unified Method" of carp capture; U.S. Geological Survey Fact Sheet 2020–3005; US Geological Survey: Columbia, MO, USA, 2020; 2p. [CrossRef]
- Sun, H.; Zhang, A.; Yan, G.; Zhang, Y.; Meng, X.; Liu, L.; Xie, N.; Cheng, W.; Wang, X. Acupuncture targeting and regulating multiple signaling pathways related to Zusanli acupoint using iTRAQ-based quantitative proteomic analysis. Acupunct. Relat. Ther. 2014, 2, 51–56. [Google Scholar] [CrossRef]
- Huang, L.; Cobessi, D.; Tung, E.Y.; Berry, E.A. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: A new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J. Mol. Biol. 2005, 351, 573–597. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Sunghwan, K.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Cupp, A.R.; Woiak, Z.; Erickson, R.A.; Amberg, J.J.; Gaikowski, M.P. Carbon dioxide as an under-ice lethal control for invasive fishes. Biol. Invasions 2017, 19, 2543–2552. [Google Scholar] [CrossRef]
- Vetter, B.J.; Murchy, K.A.; Cupp, A.R.; Amberg, J.J.; Gaikowski, M.P.; Mensinger, A.F. Acoustic deterrence of bighead carp (Hypophthalmichthys nobilis) to a broadband sound stimulus. J. Great Lakes Res. 2017, 43, 163–171. [Google Scholar] [CrossRef]
- Lennon, R.E.; Berger, B.L. A resume on field applications of antimycin A to control fish. In Investigations in Fish Control; U.S. Bureau of Sport Fisheries and Wildlife: Washington, DC, USA, 1970; pp. 1–23. [Google Scholar]
- Rach, J.J.; Luoma, J.A.; Marking, L.L. Development of an antimycin-impregnated bait for controlling common carp. N. Am. J. Fish. Manag. 1994, 14, 442–446. [Google Scholar] [CrossRef]
- Miyoshi, H.; Tokutake, N.; Imaeda, Y.; Akagi, T.; Iwamura, H. A model of antimycin A binding based on structure-activity studies of synthetic antimycin A analogues. Biochim. Et Biophys. Acta 1995, 1229, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]
- Zhao, B.; Katuwawala, A.; Oldfield, C.J.; Dunker, A.K.; Faraggi, E.; Gsponer, J.; Kloczkowski, A.; Malhis, N.; Mirdita, M.; Obradovic, Z.; et al. DescribePROT: Database of amino acid-level protein structure and function predictions. Nucleic Acids Res. 2021, 49, D298–D308. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).