Abstract
Chondrichthyes (including sharks, rays, and chimaeras) are a class of jawed cartilaginous fishes (with skeletons composed primarily of cartilage), with major relevance to the marine ecosystems and to humanity. However, cartilaginous fishes are facing various threatens, inflicting abrupt declines in their populations. Thus, critical assessment of available molecular genetic variation, particularly retrieved from Chondrichthyans’ transcriptomic analyses, represents a major resource to foster genomics research in this ancient group of vertebrate species. Briefly, RNA-Seq involves the sequencing of RNA strands present on a target tissue, which can assist genome annotation and elucidate genetic features on species without a sequenced genome. The resulting information can unravel responses of an individual to environmental changes, evolutionary processes, and support the development of biomarkers. We scrutinized more than 800 RNA-Seq entries publicly available, and reviewed more than one decade of available transcriptomic knowledge in chondrichthyans. We conclude that chondrichthyans’ transcriptomics is a subject in early development, since not all the potential of this technology has been fully explored, namely their use to prospectively preserve these endangered species. Yet, the transcriptomic database provided findings on the vertebrates’ evolution, chondrichthyans’ physiology, morphology, and their biomedical potential, a trend likely to expand further in the future.
Key Contribution:
This review aims to summarize information on the available transcriptomes resources of Chondrichthyes. Such accumulated knowledge has multidisciplinary relevance, including diverse uses on biomedical industry, inference of organ origins and the understanding of metabolic pathways.
1. Introduction
The oceans are ecosystems filled with a vast number of services to humanity, such as resources that can be collected, sequestration of carbon and consequent regulation of coastal water levels, and cultural services [1]. Unfortunately, numerous anthropogenic stressors are causing the decline of those oceans’ services, which can result in catastrophic events [2]. Chondrichthyes, a class of fishes composed by two subclasses, Chimaeras and Elasmobranchs, are crucial to the accomplishment of the oceans’ services and are particularly vulnerable to the stressors that the oceans are currently experiencing. Specifically, since Chondrichthyes are major predators, they are responsible for the top-down control of the lower trophic level [3], maintaining healthy and conventional fish stocks [4], even though some species can present different feeding behaviors such as filtering [5] and scavenging [6]. Additionally, Chondrichthyes are also involved in the carbon cycle in the oceans by feeding of the dead matter on the sea floor [7,8,9,10]. Due to their often-large size, they conserve substantial amounts of carbon in their bodies, and upon death, they sink, allowing carbon recycling when eaten by scavengers [11]. However, if they are fished, the carbon cycle is disrupted, which can compromise the worldwide climate [12]. Chondrichthyes are also highly popular due to their appearance, relevance to the movie industry, and cultural roles in some communities [13], providing valuable resources for tourism and, consequently, economically [14,15]. Nevertheless, their impact on improving the well-being of humanity can be reduced due to their populations decline, mainly caused by overfishing [16].
Understanding that biochemical changes happening on a given organism can have impacts on populations and, ultimately, ecosystems [17], calls for the development of tools that can provide such knowledge. Transcriptomic studies can deliver the necessary information that can be used for fish conservation, by researching the genetic responses to environmental stressors, and for biomarker development, used for faster stress diagnosis [18]. Such studies consist of collecting all RNA sequences, such as messenger RNA (mRNA), small RNA (smRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), and non-coding RNA [19]. The first attempts occurred in the early 1990s [20] and the development of the sequencing technologies resulted in an increasement in the transcripts’ database and their quality [21]. Indeed, transcriptome studies assist the understanding of the mechanisms and response of organisms by analyzing their genetic expression, highly relevant to identify ways of enhancing the ecologic conservation of species [18], understanding evolutionary processes and phylogenetic relationships [22] and even for disease treatment [23,24].
Here, we critically review the current knowledge and available resources about the transcriptomes of cartilaginous jawed fishes, which in line with other sequencing techniques and data, such as genomics [25,26,27], proteomics [28,29] and microbiomes [30,31,32], is likely to foster research in this fish class. We collected detailed information regarding the available sequences from the SRA NCBI Database and from literature, aiming to understand the state-of-art of Chondrichthyes’ transcriptomics. Given the huge economic and ecological potential of chondrichthyans, exploring these resources can assist humanity in many ways. For example, chimaeras’ transcriptome can help to understand the evolutionary history of vertebrates [33], while elasmobranchs have important biomedical potential [34,35] and can be used as bioindicators of marine ecosystems [36]. Furthermore, summarizing which taxonomic groups are underrepresented and which tissues are being mostly used will provide an important record of the missing data, which will allow researchers subsequently to fill possible gaps.
2. The Chondrichthyes
Chondrichthyes is a class of jawed fishes whose endoskeleton has the peculiarity of being composed by cartilage [37], and its species are separated into two different subclasses: Elasmobranchii and Holocephali. The Elasmobranchii subclass is composed of sharks (superorder Selachii), and skates and rays (superorder Batoidea), accounting for over 1200 different species of elasmobranchs [38]. On the other hand, Holocephali is composed of chimaeras, which have fewer described species than their sister taxa (elasmobranchs), with only around 56 species [38]. Elasmobranchs are represented worldwide, on the most distinct ecosystems [39], from shallow waters [40] to deep regions of more than 2400 m of depth [41], and from the cold waters of Antarctica [42] to the warm waters of tropical reefs [43]. Usually, they occupy the position of apex and mesopredators in the food chain, implicating a huge factor in the populations’ dynamic of the ecosystem they inhabit [44,45], but some species, such as whale sharks (Rhincodon typus), also present filter feeding habits [46,47]. Currently, the overfishing of Chondrichthyans is the key trigger for the extinction of many species. However, habitat destruction, climate change and pollution also contribute to this phenomenon, but on smaller proportions [48]. These anthropogenic effects influence not only population numbers but can also compromise the evolutionary potential of the affected species [49]. Additionally, the population shrinkage due to anthropogenic events can escalate due to some of their biological traits, such as late sexual maturity, lengthy pregnancy, low fertility, slow growth and long life span up to hundreds of years [50,51].
As stated above, cartilaginous jawed fishes impact our planet on countless factors. Economically, they are a valuable target for fisheries, with an estimated one billion American dollars traded owing only to shark commerce worldwide [52]. If we account for the often mislabeling of these species [53,54] or the non-reported catches [55,56], the global market of chondrichthyans can surpass the previously mentioned values, even though they are regularly thought to be bycatched [57,58]. Nevertheless, they are fished extensively on some regions [59,60,61] by industrial, artisanal, and even recreational ways [62,63]. The fished specimens can be used not only to human consumption, but also for cosmetics and pets’ food [64]. Furthermore, the potential of collagen extraction for biomaterial production is being researched for tissue regeneration [65,66] and as antitumoral [67,68], even though some claims can be debated [69]. Moreover, being such an ancestral class, it can provide key evolutionary knowledge, given the fact that there are fossils of chondrichthyans dating more than 400 million years ago [70]. In addition, this lineage survived four mass extinctions, making them one of the earliest and most successful vertebrate groups [71]. With all the evolutionary pressures and different abiotic factors they faced during millions of years, they possess some impressive features on their genome [27,34,72], such as a deletion of a whole Hox cluster, a great expansion of some vomeronasal gene-families, and positive selection of genes involved on wound-healing and genome stability. In addition, extant elasmobranchs possess low genetic diversity and a resilience that suggest unique genetic properties that must be uncovered and preserved [73]. Consequently, analyzing their transcriptomes will be relevant to understanding the genes that are expressed and under what circumstances.
3. Evolution of Transcriptomics Technologies
Transcriptome studies have undergone a huge development since the beginning of sequencing practices, with the emergence of innovative techniques that bring outcomes with greater accuracy, less need for computational tools [74,75], and therefore, faster results [76]. The usage of RNA for omics highly increases the difficulty of sequencing due to its single-stranded structure, which is very unstable and can compromise the sequencing results [77]. Thus, it is fundamental to use proper reagents that can preserve the samples (e.g., RNA Later), store the samples at lower temperatures, or both, depending on the time between sampling and extraction [78,79]. Nevertheless, the first sequencers were only capable of sequencing DNA, therefore efforts of understanding the transcriptome of an organism appeared around the 1970s with the usage of reverse transcriptase and cDNA amplification by PCR, followed by sequencing [80]. The main limitation of this generation was the reduced amounts of DNA that could be processed and the initial high costs [81].
The following generation, named NGS (Next Generation Sequencing), was established on the mid 2000s providing faster results, which overtime evolved to more welcoming costs [82] for a higher performance rate than the first generation [83]. These technologies were based on high throughput sequencing (HTS). With this generation, transcriptomics had an increasing attention, since one of the advantages is the possibility of quantifying transcripts and following comparisons [84]. Succinctly, transcriptome analysis using NGS, can be divided into three major steps [85]: Library construction, consisting of the selection and collection of the appropriate samples, following by RNA extraction, reverse transcription, and cDNA fragmentation (the last two steps can be performed on different order) [86]. The next step is the sequencing itself, which varies on the technology used [87]. Finally, bioinformatic analyses of the many short sequences obtained, will be distinctly targeted depending on the different focus of the carried-out research [88,89].
Additionally, it is important to mention the microarray technology, which was one of the initial drivers of the gene expression research. Microarrays were used to evaluate genetic expression, in the earlier moments of sequencing development, and it was a cheaper option, requiring less computational power [90] Even though this technique is still in use [91,92], the need of having previous molecular knowledge on the targeted species, limited the species compatible with this technique [93,94], and the NGS development led to the reduced interest of microarrays. Nevertheless, they can still be relevant to evaluate gene expression in model species [95,96,97].
Currently, the latest generation of sequencing is being developed, which focus on longer single reads that can reach values of 150 kbp [98] and the ability of direct sequencing RNA [99] and not need amplification before sequencing [81]. These long reads diminish the complexity of the assembling process, and can improve genome annotations [100], de novo assembly [101], and epigenome characterization [102]. This technology has the huge advantage of not needing amplification via PCR [103], which can reduce the PCR-related mutation errors, labor and reagents usability. Nevertheless, the earlier times of this generation brought a higher error rate that is currently being diminished [104,105], and it can be further reduced using hybrid techniques, long read sequencing combined with short reads for refining assemblies [106].
4. Chondrichthyes and Their Transcriptomes
We assessed the available transcriptomes in the literature and databases, utilizing Web of Science, with a combination of keywords (sharks, skates, rays, chimaeras, elasmobranchs, chondrichthyes, transcriptome, transcriptomics, gene expression). We analyzed this information along with all NCBI SRA database entries for RNA, filtering for Chondrichthyes species from January 2011 until, 10 April 2023. Our data revision shows that the number of RNA-Seq entries have been increasing since 2019, and with two years of higher number of entries (2018 and 2022, with 218 and 253 entries, respectively), a trend that can be seen on Figure 1. Regarding the early months of 2023, there were already 55 new entries. The boom of entries occurring in 2018 is mostly related with the work by Hara et al. [107] with more than 100 entries, and Swenson et al. [108] which accounts for more than 50 entries. Likewise, the year of 2022 had the work published by Mayeur et al. [109] that contributed solely with almost 100 entries. When researching the transcriptomes available on NCBI platform [110], using the SRA database, filtered for RNA entries and for bony fishes and cartilaginous fishes, it is clear the discrepancy between these two groups. Bony fishes account for more than 90,000 entries, whereas Chondrichthyes have under 900 entries. From all these entries, if we excluded scientific replicas and repeated tissues per species, there are only 198 unique tissues sequenced from Chondrichthyes’ species, which were divided by different categories following the tissue origin (Figure 2). The most sampled tissue for transcriptome studies on Chondrichthyes is the muscle, which can be explained by the easy sampling and stable preservation. Moreover, most tissues require the sacrifice of the individual. Given their conservation status, it would be optimal to sequence less invasive tissues which are proven as a viable method on wild fishes [111].
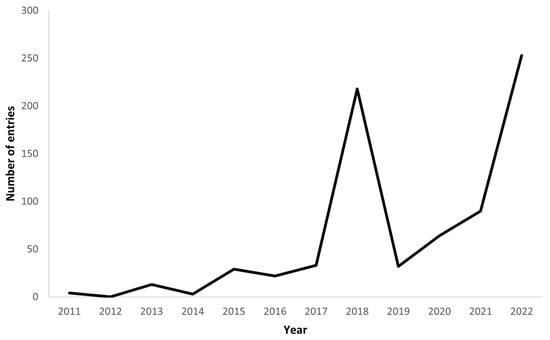
Figure 1.
Chondrichthyes’ RNA entries available in the public dataset from 2011 to 2022.
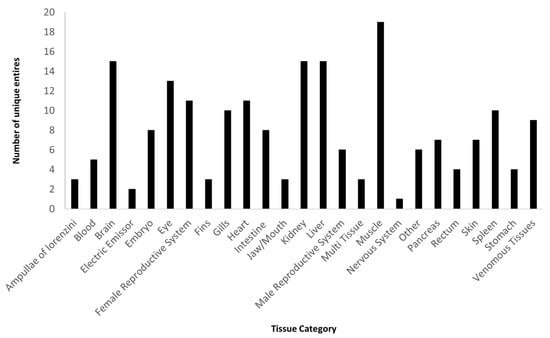
Figure 2.
Number of unique categorized RNA sequences available by tissue for different Chondrichthyes species.
Regarding their conservation status, Figure 3 shows the entries by the species’ IUCN Red List Status, divided by their orders. Most of the entries belong to Least Concern species (64.2%), whereas the least represented belongs to Critically endangered species (1.7%), which can be related due to their lesser abundance on ecosystems (excluding unknown status). Notwithstanding, more effort should be placed on sequencing these less abundant and more endangered species, since it could improve their conservation based on a better understanding of their biochemical genetics. Moreover, from all the species known of Chondrichthyes, only 54 species (3.7 %) had their transcriptome studied, and from those only 1 species (0.07%) belongs to the Chimaeras, 29 species (1.4%) to the Batoidea, and 33 species (2.2%) to Selachii. This highlights the importance of increasing the sampling of chimaeras’ tissues, since these lineages of elasmobranchs and chimaeras diverged 375 million years ago [112].
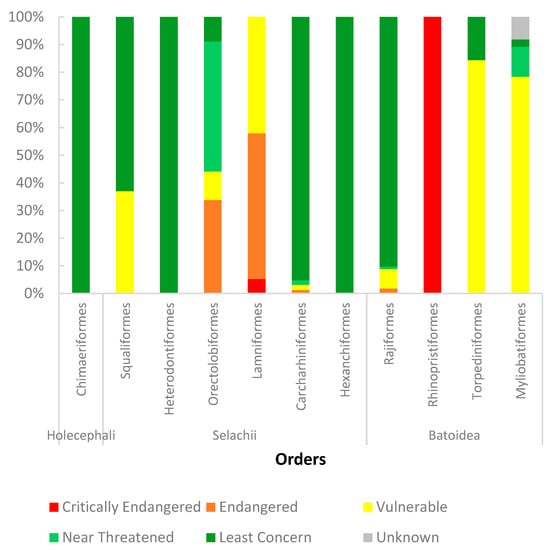
Figure 3.
Percentage of IUCN Red List Status species represented with entries in each order.
When evaluating the sequenced tissues by orders, the chimaera C. milii solely represents the only Chimaeriformes order. Regarding the Batoidea infraclass that comprises four orders (Myliobatiformes, Rhinopristiformes, Rajiformes, Torpediniformes), there is at least one species sequenced for each of the existent order. By contrast, Selachii infraclass has eight orders (Carcharhiniformes, Heterodontiformes, Hexanchiformes Lamniformes, Orectolobiformes, Pristiophoriformes, Squaliformes and Squatiniformes), and not all are represented with a transcriptome entry. Regarding the diversity of tissues sampled for each species (Figure 4), only seven species have 10 or more tissues sampled: Scyliorhinus torazame, Callorhinchus milii, Stegostoma fasciatum, Chiloscyllium punctatum, Rhincodon typus, Scyliorhinus canicula, and Leucoraja erinacea [113,114,115,116,117]. Moreover, only one species besides Leucoraja erinacea on the Batoidea infraclass had 10 or more different tissues sampled, whereas Narke japonica had the most tissues sampled [9].
Given the importance of transcriptomics for genome annotation [118] and the evolution of the sequencing technologies, it is expected that more species will have both their genome and transciptome sequenced. However, only 17 species currently have their genome sequenced on NCBI Databases, which accounts for only 1% of the known Chondrichthyes species. Consequently, there are various orders without whole-genome data available, and efforts to increase the genetic knowledge of these animals should be a prime concern for future sequencing projects [27].
Thus, it is important to gather the information available on the transcriptome and gene expression of elasmobranchs and chimaeras, and then take advantage of the new available technologies to complete the databases. The usage of the long-read sequencing in Chondrichthyes is still uncommon, but besides the biological information it can deliver, it can also be viable for facilitating the genome annotation on some of the sequenced species [119] due to their long reads, easier assembly, and to perform isoform identification [120]. Even though previous generations of transcriptomics could help the annotation of genomes [107], their performance is less effective to decipher highly repetitive parts of the genome [121]. Even with such a low sampling number, it is possible to explore how transcriptome data can help understand the evolution of vertebrates, the physiology, and morphology of these species, and how these animals can help to enhance the human’s well-being.
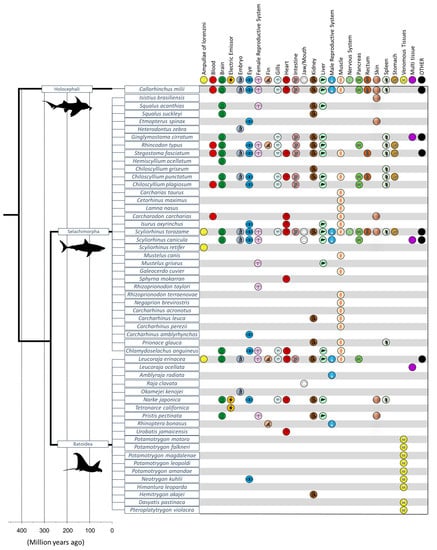
Figure 4.
Taxonomic distribution of chondrichthyan’s transcriptomic entries in the NCBI database. Organized by species and organs where the tissues were sampled. The category “OTHER” includes tissues, such as epigonal, gallbladder, head sections, olfactory epithelium, axial skeleton sting epithelium, saccus vasculosus, thymus, and nonidentified tissues). Divergence data based on Timetree [122].
5. Main Findings of Elasmobranch’s Transcriptome Studies
The first transcriptomic study on cartilaginous fishes [123] was reported in early 2011, aiming to better understand the genetic changes of vertebrates’ evolution and the uniqueness of elasmobranchs genetics. Embryos from Scyliorhinus torazame were sequenced using Sanger and NGS methodologies to obtain expressed sequence tags (ESTs) that revealed the presence of genes involved on jaw patterning, absent on hagfishes, a jawless fish. Two years later, in 2013, the transcripts of the heart of the great white shark, Carcharodon carcharias, were sequenced, but the lack of content on public databases hindered the gene annotation of its transcripts. Nevertheless, many Genetic Ontology terms related to humans and zebra fishes were annotated, and surprisingly, C. carcharias had more similarities with humans than with Danio rerio [124].
5.1. Evolutionary Findings
5.1.1. Pancreas
Due to their evolutionary importance, and with the increasing availability and decreasing costs of sequencing processes, more studies were performed to gather cues to decode vertebrates’ evolution. For instance, studies regarding pancreatic emergence were performed on a dogfish, since jawless hagfish and lampreys do not possess a defined pancreas, but Chondrichthyes present what can be qualified as an “ancient pancreas” [125]. Surprisingly, when comparing the transcriptome of the liver, pancreas, and brain, there are more similarities between the pancreas and the brain, than the pancreas and liver, even though they have more similar functions [126]. These results can explain the similar origin of neural and pancreatic endocrine cells [127].
5.1.2. Eyes
Tissues from the same region can be compared, not only between Chondrichthyes, but also between other species. Given their evolutionary position, phylogenetically located between jawless vertebrates and bony fishes [128], comparative studies between these groups can elucidate some of their evolutionary histories. With the objective of clarifying the evolution of the vertebrate phototransduction cascade, RNA from the eyes of hagfish—Eptatretus cirrhatus, lampreys—Geotria australis and Mordacia mordax, elasmobranchs—Chiloscyllium punctatum, Carcharhinus amblyrhynchos, and Neotrygon kuhlii, bony fish—Amia calva, and gar—Lepisosteus platyrhincus [129] were sequenced and revealed that elasmobranchs have similar mechanisms of phototransduction cascade as the bony fishes, using GNAT1 together with PDE6, unlike, agnathostome that only has GNAT1. Regarding Isurus oxyrinchus, the transcriptomes of the eye were analyzed and there was an overexpression of the photoreceptor CRB1, which suggests that they utilize vision as their primary sense, and the expressed ocular opsin genes revealed a monochromatic vision on this species [130]. Furthermore, it has been suggested in S. canicular the reduction of growth in the retina due to a decreased expression of genes typically associated with cellular development and growth [131].
5.1.3. Heart
Additionally, when comparing the heart transcriptomes of seven species (four elasmobranchs and three teleosts) it was estimated that half of the transcribed genes were present in all the species. Some of the differences appeared in immune functions, with two or more of the studied elasmobranchs possessing 30 of the 37 genes related to adaptive immunity genes which are absent from the researched teleosts and C. milii’s genome [132], reinforcing the importance of evolutionary research on the subgroup of elasmobranchs.
5.1.4. Reproduction
Differences in the reproductive strategies of different shark species were explained by RNA-Sequencing the white muscle from species with placental development (Rhizoprionodon terraenovae, Carcharhinus acronotus, Prionace glauca, Carcharhinus leucas, Carcharhinus perezii, Mustelus canis and Negaprion brevirostris), and non-placental development (Carcharias taurus and Galeocerdo cuvier). These authors point out that signal of positive selection in genes associated with brain development (YWHAE and ARL6IP5) and sperm production and morphology (TCTEX1D2 and VAMP4) can be influencing the placenta loss on Carcharias taurus and Galeocerdo cuvier [133].
5.1.5. Metabolism
Regarding an ancient representative of cartilaginous fishes, Callorhinchus milii, a chimaera, one study comparing the transcripts from different tissues showed unexpected gene expression on myoglobin (Mb), α-globin, and globin X (GbX) genes [134]. Detailing these differences, Mb showed a higher expression in the heart than on skeletal muscle, hypothesizing that higher aerobic metabolic rates evolved to Mb being expressed on skeletal muscle to increase oxygen demands. Moreover, both α-globin (α1) and β-globin showed higher expression in the heart and spleen, which was expected for subunit isoforms of a tetrameric hemoglobin within red blood cells, but the paralog α2 was most expressed in the brain. The authors hypothesize that the lack of the NgB gene is replaced by α-globin (α2). Likewise, the paralog of GbX1 was expressed on a diverse range of tissues, while GbX2 was highly expressed on the gonads. Furthermore, when comparing the transcriptome of the white muscle of three shark species with six tuna species and a mackerel, it was possible to hypothesize that the gene glycogenin-1 relates to the fast recovery of exercise and can be associated with the evolution of endothermy in sharks and tunas [135]. Moreover, transcriptome analysis of the Sphyrna zygaena’s liver showed the possibility of lectin pathway being gone in the hammerhead shark lineage, although it was supposedly established on a common ancestor of bony fishes and cartilaginous fishes [136].
5.2. Physiology and Morphology
5.2.1. Fin Development
The morphology of the cartilaginous fishes’ body is also an interesting topic of study, since some Batoids possess a pectoral fin that fuses with the head [137], which can be explained by the different genetic expression on the posterior region of pectoral fins versus the anterior region [138]. Moreover, transcriptomics was used as a comparison method to understand how rays from the family Myliobatidae developed “horns”, formally called cephalic lobes, that allow them to inhabit pelagic environments [139]. When comparing the pectoral fins’ transcripts of L. erinacea with Rhinoptera bonasus, the genes Alx1, Alx4, Pax9, Hoxa13, Hoxa2, and Hoxd4 were the most common in both species. On the other hand, it was found that the genes Dkk1, Msx2, Omd, and Lhx2 are possible inducers of the formation of the notch on Myliobatidae, and there was a downregulation of Msx1 (which is associated with a proper maturation of apical ectodermal ridge, on the notch development) and Bco2 (boosts endogenous retinoic acid synthesis) [108]. The development of sequencing technologies turned them cheaper and faster over time, opening the possibility of producing a tridimensional image of the RNA Profiling on zebrafish, combining techniques of RNA-Seq and computed tomography [140]. A similar methodology was used on S. canicula, on the forebrain of the embryos [109], even though the resolution was not at a cellular level, as the third-generation sequencing enables. It was possible to correctly show the expression details, such as the differences of the paralogs of Nkx, Six, and the asymmetric left/dorsal restrictors, Lefty2/Nodal/Vg1. This study showed that application on a larger organism was possible (similar methodologies are usually utilized on smaller species [141]) and help decipher the ancestral properties of jawed vertebrates. Transcriptomic studies can also be valuable to understand the genetic expression of motoneurons and the evolution of tasks like land walking. This was shown, for instance, with the identification of transcription factors in Leucoraja erinacea embryos that are identified in mice as responsible for the specification of motoneurons—e.g., Foxp1. Additionally, the Leucoraja fins express the same genes as the lateral motor columns (LMC) of mice and chicks, and the ray expansion of LMC is due to the absence of the Hoxc9 gene [142]. Likewise, to understand the genetic changes in the development of fins and limbs, pectoral fins from C. punctatum and forelimb buds from mice were sequenced at different times of development. The study revealed that even though many of the expressed genes were similar in both species, their activity was different. Some genes in mice were expressed in the late stages of the member development, whereas the same genes were shut down at the same stage of fin development [143].
5.2.2. Digestive System
Nutrient uptake can also be studied with the usage of transcriptomic techniques. Honda et al. [144] sampled embryonic intestine and yolk sac membrane from S. torazame and realized that four months before hatching there was an increase of many amino acid transporter genes, and lipid absorption in the intestine. About the yolk sac membrane, the increase in basolateral amino acid transporters and cathepsin in late development stages was reported. The authors hypothesized that the amino acids and lipids from the yolk are transported through the basolateral membrane into the blood. Some interesting physiological aspects of these creatures were molecularly explained with the help of transcriptome studies, such as the functional processes involved in the rectal gland, which has an important role in the osmoregulation processes but is also connected with the feeding process [145]. In Squalus acanthias’s case, this mechanism was supported by the storage of crucial mRNAs that will trigger a faster translation of proteins, when the gland is activated by feeding [146].
5.2.3. Osmoregulation
Recurring once again to the S. acanthias’ transcriptome, multiple tissues were sequenced (brain, liver, kidney, and ovary) with a key focus on understanding the osmoregulation processes. The presence of urea synthesis genes in the liver was clear, even though some studies also reported it on other tissues [147]. Moreover, when studying the transcripts of S. acanthias the presence of two glutamine synthetase (Gs) orthologues was identified, which were not detected in previous studies [148]. Regarding arginase, an important enzyme involved in the urea cycle, known for its versatility [149], two orthologs (Arg1, Arg2) were detected. While Arg1 was only detected on the kidney assembly, Arg2 was found on all the four sequenced tissues. Lastly, aquaporins (AQP) were also detected, which are involved in urea reabsorption processes [150] (a process that is still not well understood). It was notable the presence of two Aqp3 genes that can help increase knowledge on this gene subclass. Remarkably, some shark species are capable of occupying fresh water and saline environments, such as the bull shark Carcharhinus leucas [151]. Therefore, in order to understand this feature, kidneys of bull sharks acclimated to freshwater and saltwater were RNA-sequenced. It was expected that the NKCC2 was responsible for the reabsorption of NaCl when transitioning from seawater to freshwater, but there were no differences detected in the gene expression of both kidneys. In contrast, an increase in the expression of Na+-Cl− co-transporters and Na+/K+ ATPase subunit α1 (NKAα1) was noticed [152]. NKAα1 is responsible for generating Na+ electrochemical gradient, which is the driver of NaCl reabsorption [153]. Similarly, Hemitrygon akajei, demonstrated a co-expression of the genes NKCC2 and NKAα1 in the tubular bundle of the kidney, while dealing with lower salinity environments [154].
5.2.4. Climatic Adaptation
Given the climate changes that ecosystems are currently facing, it is important to identify the variations that can happen in elasmobranchs’ genetic features. For example, according to Lighten et al., Leucoraja ocellata could adapt to higher temperatures majorly by reducing their body size, but also due to changes in their genetic expression [155]. Curiously, there were also changes in genes involved in immune response, hypothesized as a response to the secondary effects of thermal stressors, which is a reported effect on some species [156].
5.2.5. Electric Organs
Furthermore, some Chondrichthyes have unusual features, such as Tetronarce californica, a ray with the capability of emitting electric shocks with hundreds of volts [157]. Transcriptome analysis of the electric lobes of this species concluded that the electric shocks are the outcome of a synchronized neurotransmitter-mediated depolarization, with the involvement of genes that encode the Excitatory Amino Acid Transporter 1 (EAAT1), Chloride Channel protein 2 (ClC-2) and Voltage-dependent L-type calcium channel subunit 1 (Cav1.2). Furthermore, Dispanin and V-type proton ATPase 16 kDa proteolipid subunit were detected, and both these proteins are present in other electric rays [158]. Additionally, some species are not only able to discharge electric shocks, but also of sensing electric signals by using the organ Ampullae of Lorenzini [159]. This organ was sequenced on L. erinacea for their transcriptome. The results showed that high conductance might result from highly expressed calcium-activated potassium (BK) channel and voltage-gated calcium channel (CaV1.3) on receptor cells (but not on the support cells or tubule structures). These channels can be mediated by the most expressed gene parvalbumin-8 [160].
5.2.6. Bioluminescence
The deep-sea shark, Etmopterus spinax, which is capable of emitting a blue-green ventral glow [161], had his eyes and ventral skin sequenced for transcriptome analysis. The transcripts for Es-rhodopsin and Es-peropsin were solely detected on the eye, confirming the monochromatic vision of this species. In contrast, Es-encephalopsin was found on both tissues, but more on the ventral skin, and it is hypothesized that it initiates pathways leading to ultraviolet radiation phototransduction on the skin [162]. Similarly, Isistius brasiliensis also presents bioluminescence capabilities [163]. Ventral tissues (bioluminescent part of the body) and black band integument were sampled, and their transcripts were sequenced. However, it was not possible to find similarities of any of the identified genes with the conventional bioluminescence enzyme present in insects, luciferase [164]. However, there were 30 genes that were only expressed in the bioluminescence part of the body, which can bottleneck the investigation of the mechanism involved in bioluminescence in these sharks [165].
5.2.7. Anoxia Response
While sequencing Hemiscyllium ocellatum’s brain, and comparing it with a frog, a carp, and a turtle, it was noticed that small ncRNA were differently expressed while exposed to anoxia, while on the following recovery phase accounted for less than 1% in each studied species. In the shark’s transcripts, the majority was not possible to annotate as a known RNA [166]. Some of these can be expressed for the immediate effects of anoxia, and others for the survival of long-term survival of anoxia. More studies can help clarify the functions of each of those small ncRNA.
5.3. Biomedical Relevance
5.3.1. Venom
While venoms are frequently harmful and can be deadly in some cases [167], they are also promising for their biomedical potential on many diseases [168]. Some ray species possess venom glands that can be used for transcriptome studies, such as Potamotrygon amandae, Potamotrygon falkneri and Potamotrygon motoro, freshwater species that possess a non-lethal venom. The venom glands’ transcriptome showed, as expected, numerous proteins related to envenomation processes [169,170], such as thrombin-like enzymes, that can form thrombus [171]; hyaluronidase, which improves the diffusion of fluids across the skin, enhancing the venom potential [172]; phospholipases, which are quoted as one of the most dangerous toxins on animals, possessing a large spectrum of activity [173], and proteinases, specially metalloproteinases [174]; glycoproteins such as CRISPs, which are hypothesized to disorder the homeostasis of the infected organisms [175,176]; and neurotoxins, for example ohanin and α-atrotoxin-Lt1a [177,178]. This knowledge can be important not only for the biology of the species and for the discovery of novel proteins, but it can also help clinical diagnosis when a patient is envenomated. Regarding the marine ecosystems, Neotrygon kuhlii was studied for its transcriptome, in combination with proteomics, showing that galectin was highly expressed and was abundant on the venom proteome [179]. This protein is known for changing blood dynamics [180]. Furthermore, peroxiredoxin-6 was highly expressed too, which is curious since it works as an antioxidant, but in some metabolic pathways can lead to toxic activities. Kirchhoff et al. (2022) [181] performed a study showing the capabilities of transcriptomics for the search of those proteins with biotechnological potential. Combining the transcriptome sequencing of five ray species (Potamotrygon leopoldi, Potamotrygon motoro, Dasyatis pastinaca, Himantura leoparda and Pteroplatytrygon violacea) with Hella cell bioactivity assays (LOPAC 1280 library) in a network, they could indicate 216 signaling pathways, where 29 of those were shared by 70 transcripts and 70 bioactivity hits. This methodology can help drug discovery of other unusual venomous species.
5.3.2. Kidney and Spleen
It is thought that sharks were the ancestral group displaying innate and adaptive immune system [182]. Chiloscyllium griseum’s spleen and kidney were RNA-sequenced and their spleen showed immune and signaling pathways, with cell adhesion molecules and various receptors, whereas the kidney showed more gene expression on metabolic pathways, such as xenobiotic degradation and lipid, amino acid, and carbohydrate metabolism [183].
5.3.3. Antibodies
Moreover, Chiloscyllium plagiosum is a focus species in some scientific fields, for presenting some appealing compounds [184], being used as model species on ecotoxicological studies [185] and for the interest in their single domain antibodies [186]. The latter is being studied on a production level, and the transcriptome was sequenced on different tissues of this species [187]. It was possible to understand that IgNARs antibodies were produced mostly on the spiral valve, pancreas, and spleen. IgNAR1 was mostly expressed in the adult stages, but IgNAR6 in juveniles. These discoveries can help increase of efficiency of the production of these antibodies, depending on the interest of each antibody. Moreover, the usage of multi-tissue transcriptomics aided with the identification of more than 626 nurse shark plasma proteins [188], which helped identifying the proteins involved on the response of immune stressors on Ginglymostoma cirratum. This can be important for understanding how chondrichthyes respond to immune stimulants and help with the production of antibodies.
5.3.4. Liver
The frequently sequenced C. plagiosum species was also used for transcriptomic studies due to its unusual liver that occupies more than 70% of its visceral weight. Studies in this species’ liver have reported molecules with potential in the biomedical field, which can make C. plagiosum a possible model species to evaluate liver regeneration [189,190]. Therefore, the importance of microRNAs (small, non-coding, single-stranded RNAs, 19 to 25 nucleotides in length, which regulate biological processes [191]) on the regulation of liver regeneration was researched in this species, showing the expression of microRNAs related to liver regeneration mostly 3–12 h after partial hepatectomy, with more than 300 differentially expressed microRNAs [192]. Studies like this can help the research of bioactive compounds, and elucidate their mechanism of action. Moreover, peptides from C. plagiosum were already tested for their potential for liver regeneration studies [193]. Regarding Isurus oxyrinchus, the transcriptomes of the liver, showed that most of the genes expressed on this tissue had functions related to resistance to different cancers and other human diseases, such as HABP2 and PONs’ genes [130].
6. Challenges and Future Perspectives
As it was noted on this review, the transcriptome analysis of the Chondrichthyes’ tissues is still a very understudied subject, following the same pattern as the genomic studies regarding the same class of organisms [27]. With so many astonishing features that these species possess, there are numerous aspects that can be studied, such as elucidating the development of the large tail of thresher shark [194], understanding how some species possess faster and more efficient reproduction strategies, namely P. glauca [195] and S. canicula [196], or even which are the biochemical regulators behind the different morphologies of the various species of hammerhead sharks [197].
The lack of data on the Chondrichthyes’ transcriptomes is a combination of many factors, beginning with the logistics of RNA preservation that can be challenging and costly on field work [198]; chondrichthyes’ meat presents a low commercial value (even though fin market is highly valuable) [199] which can lower the economic interest of preserving this class; and the difficulty of sampling these specimens [200] due to their biology and costs of marine expeditions. To tackle this, opportunistic sampling can be performed while on board of commercial or recreational fishing boats, as previously accomplished [201,202,203]. This can limit the experimental design due to the inconsistent sampling. Nevertheless, it is remarkable how the possibility of maintaining some species in captivity can help better understand the genes involved in the characterization and metabolism of each of these species, using their embryos [144,204], and changing different settings to evaluate how some species cope with different stressors [166]. With the increasing number of studies regarding the pollutants present on elasmobranchs, it is important to understand how these organisms are coping with such high levels of pollutants [205,206,207], which was proved as a good possibility of study using biomarkers [208]. Likewise, the effects of climate changes and following repercussions (such as ocean acidification) are thoroughly explored [185], but currently lack studies at genetic expression level. Comparing the differential genetic expressions and functional analysis between the presence and absence of various stressors can decipher which factors are influencing the fitness of the individuals and understand sub-lethal limits [209]. It would also benefit the development of novel and more specific biomarkers [210] and support the creation of supportive laws to enhance the cartilaginous fishes’ conservation [18]. Nevertheless, transcriptomic studies are not a universal tool, and it should be used along with other analyses to provide a correct image of the fitness of the target and its protein activity [211]. Additionally, robust comparisons should be established between lethally-sampled tissues such as kidney, liver, and brain (well known for their role in homeostasis, detoxification, and nervous system control, respectively), and tissues that can be sampled using non-lethal strategies [111], with the objective of evaluating gene expression on a tissue and correlate with a response of different organs. Non-lethal tissue sampling can enhance the possibility of resampling the same individual over time [212], reducing the need to sacrifice individuals [91] and can lead to establishing protocols that can combine, for example, tagging animals and blood sampling or biopsy muscle punches [213]. Regarding population genetic studies focusing on cartilaginous fishes, transcriptome data is not yet commonly used, but have been applied for invertebrates, to determine how different populations are coping in different habitats [214]. This can be used on Chondrichthyes’ populations inhabiting different regions to assess putative adaptive responses to local abiotic and biotic factors. Even though that is an uncommon practice, RNA-Seq can also be used for mitogenome assembly using bioinformatic tools [215]. This can subsequently be used for population structure analysis [216]. Regarding inbreeding rates, which is an important aspect when considering populational studies, transcriptomic data may not be the most suitable tool to evaluate such parameter. Nonetheless, the effects of higher inbreeding rates have been researched on other species, with the objective of understanding how inbreeding affects the genetic expression [217].
In the future, more datasets should be provided, which can help decode the transcriptome of these cartilaginous fishes. The lack of available genomes reduces the accuracy of the transcriptome mapping since most species are not sequenced at genome level, which can difficult the interpretation of RNA-Seq results. Moreover, the usage of long read sequencing techniques is still reduced on these organisms. The advantage of in situ sequencing can lead to a diminishing need of reagents for RNA preservation and refrigeration equipment for conserving samples until sequencing. Although the cost of this kind of technology is considered high, it is expected that it will continue to reduce, providing many resources that have already showed remarkable potential. Moreover, it can provide the capability of community usage by accessibility from public databases, which can increase the knowledge and interpretation of data, after the authors publish their datasets.
Author Contributions
M.J.S.: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing—original draft, Writing—review and editing. R.R.D.: Conceptualization, Supervision, Validation, Visualization, Writing—review and editing. A.A.: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
M.J.S. was supported by the BYTPhD program through the strategic funding UI/BD/151400/2021 through national funds provided by Fundação para a Ciência e Tecnologia (FCT). R.R.D. was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (#2019/10201-0; #2022/05068-1). A.A. was partially supported by the Strategic Funding UIDB/04423/2020 and UIDP/04423/2020 through national funds provided by FCT and the European Regional Development Fund (ERDF) in the framework of the program PT2020, and by the projects PTDC/CTA-AMB/31774/2017 (POCI-01-0145-FEDER/031774/2017), Ocean3R (NORTE-01-0145-FEDER-000064), and Atlantida (NORTE-01-0145- FEDER-000040).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data will be made available upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Teneva, L.; Strong, A.L.; Agostini, V.; Bagstad, K.J.; Drakou, E.G.; Ancona, Z.; Gjerde, K.; Hume, A.C.; Jickling, N. Estimating the Pelagic Ocean’s Benefits to Humanity Can Enhance Ocean Governance. Mar. Policy 2022, 136, 104906. [Google Scholar] [CrossRef]
- Sandifer, P.A.; Sutton-Grier, A.E. Connecting Stressors, Ocean Ecosystem Services, and Human Health. Nat. Resour. Forum 2014, 38, 157–167. [Google Scholar] [CrossRef]
- Heithaus, M.; Frid, A.; Vaudo, J.; Worm, B.; Wirsing, A. Unraveling the Ecological Importance of Elasmobranchs. In Sharks and Their Relatives II; CRC Press: Boca Raton, FL, USA, 2010; pp. 611–637. [Google Scholar] [CrossRef]
- Baum, J.K.; Worm, B. Cascading Top-down Effects of Changing Oceanic Predator Abundances. J. Anim. Ecol. 2009, 78, 699–714. [Google Scholar] [CrossRef] [PubMed]
- Abreo, N.A.S.; Blatchley, D.; Superio, M.D. Stranded Whale Shark (Rhincodon typus) Reveals Vulnerability of Filter-Feeding Elasmobranchs to Marine Litter in the Philippines. Mar. Pollut. Bull. 2019, 141, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Heithaus, M.R. Predator-Prey and Competitive Interactions between Sharks (order Selachii) and Dolphins (suborder Odontoceti): A Review. J. Zool. 2001, 253, 53–68. [Google Scholar] [CrossRef]
- Tucker, J.P.; Vercoe, B.; Santos, I.R.; Dujmovic, M.; Butcher, P.A. Whale Carcass Scavenging by Sharks. Glob. Ecol. Conserv. 2019, 19, e00655. [Google Scholar] [CrossRef]
- Lea, J.S.E.; Daly, R.; Leon, C.; Daly, C.A.K.; Clarke, C.R. Life after Death: Behaviour of Multiple Shark Species Scavenging a Whale Carcass. Mar. Freshw. Res. 2019, 70, 302–306. [Google Scholar] [CrossRef]
- Roff, G.; Doropoulos, C.; Rogers, A.; Bozec, Y.M.; Krueck, N.C.; Aurellado, E.; Priest, M.; Birrell, C.; Mumby, P.J. The Ecological Role of Sharks on Coral Reefs. Trends Ecol. Evol. 2016, 31, 395–407. [Google Scholar] [CrossRef]
- NOAA Office of Ocean Exploration and Research. Dive 07: Oh My Grouper, Look at That Shark: Windows to the Deep 2019: Exploration of the Deep-Sea Habitats of the Southeastern United States. Available online: https://oceanexplorer.noaa.gov/okeanos/explorations/ex1903/dailyupdates/june28/media/sharks-log.html (accessed on 5 May 2022).
- Higgs, N.D.; Gates, A.R.; Jones, D.O.B. Fish Food in the Deep Sea: Revisiting the Role of Large Food-Falls. PLoS ONE 2014, 9, e96016. [Google Scholar] [CrossRef]
- Mariani, G.; Cheung, W.W.L.; Lyet, A.; Sala, E.; Mayorga, J.; Velez, L.; Gaines, S.D.; Dejean, T.; Troussellier, M.; Mouillot, D. Let More Big Fish Sink: Fisheries Prevent Blue Carbon Sequestration-Half in Unprofitable Areas. Sci. Adv. 2020, 6, eabb4848. [Google Scholar] [CrossRef]
- Skubel, R.A.; Shriver-Rice, M.; Maranto, G.M. Introducing Relational Values as a Tool for Shark Conservation, Science, and Management. Front. Mar. Sci. 2019, 6, 53. [Google Scholar] [CrossRef]
- Gallagher, A.J.; Huveneers, C.P.M. Emerging Challenges to Shark-Diving Tourism. Mar. Policy 2018, 96, 9–12. [Google Scholar] [CrossRef]
- Gallagher, A.J.; Hammerschlag, N. Global Shark Currency: The Distribution Frequency and Economic Value of Shark Ecotourism. Curr. Issues Tour. 2011, 14, 797–812. [Google Scholar] [CrossRef]
- Pacoureau, N.; Rigby, C.L.; Kyne, P.M.; Sherley, R.B.; Winker, H.; Carlson, J.K.; Fordham, S.V.; Barreto, R.; Fernando, D.; Francis, M.P.; et al. Half a Century of Global Decline in Oceanic Sharks and Rays. Nature 2021, 589, 567–571. [Google Scholar] [CrossRef]
- Lemos, M.F.L. Biomarker Studies in Stress Biology: From the Gene to Population, from the Organism to the Application. Biology 2021, 10, 1340. [Google Scholar] [CrossRef] [PubMed]
- Connon, R.E.; Jeffries, K.M.; Komoroske, L.M.; Todgham, A.E.; Fangue, N.A. The Utility of Transcriptomics in Fish Conservation. J. Exp. Biol. 2018, 221, jeb148833. [Google Scholar] [CrossRef]
- Morozova, O.; Hirst, M.; Marra, M.A. Applications of New Sequencing Technologies for Transcriptome Analysis. Annu. Rev. Genom. Hum. Genet. 2009, 10, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics Technologies. PLoS Comput. Biol. 2017, 13, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA Sequencing: The Teenage Years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Wen, J.; Egan, A.N.; Dikow, R.B.; Zimmer, E.A. Utility of Transcriptome Sequencing for Phylogenetic Inference and Character Evolution. In Next-Generation Sequencing in Plant Systematics; International Association for Plant Taxonomy (IAPT): Stockholm, Sweden, 2015; pp. 1–42. [Google Scholar] [CrossRef]
- Supplitt, S.; Karpinski, P.; Sasiadek, M.; Laczmanska, I. Current Achievements and Applications of Transcriptomics in Personalized Cancer Medicine. Int. J. Mol. Sci. 2021, 22, 1422. [Google Scholar] [CrossRef]
- Oliverio, A.L.; Bellomo, T.; Mariani, L.H. Evolving Clinical Applications of Tissue Transcriptomics in Kidney Disease. Front. Pediatr. 2019, 7, 306. [Google Scholar] [CrossRef]
- Domingues, R.R.; Bunholi, I.V.; Pinhal, D.; Antunes, A.; Mendonça, F.F. From Molecule to Conservation: DNA-Based Methods to Overcome Frontiers in the Shark and Ray Fin Trade. Conserv. Genet. Resour. 2021, 13, 231–247. [Google Scholar] [CrossRef]
- Johri, S.; Doane, M.P.; Allen, L.; Dinsdale, E.A. Taking Advantage of the Genomics Revolution for Monitoring and Conservation of Chondrichthyan Populations. Diversity 2019, 11, 49. [Google Scholar] [CrossRef]
- Pearce, J.; Fraser, M.W.; Sequeira, A.M.M.M.; Kaur, P. State of Shark and Ray Genomics in an Era of Extinction. Front. Mar. Sci. 2021, 8, 744986. [Google Scholar] [CrossRef]
- Lee, J.; Valkova, N.; White, M.P.; Kültz, D. Proteomic Identification of Processes and Pathways Characteristic of Osmoregulatory Tissues in Spiny Dogfish Shark (Squalus acanthias). Comp. Biochem. Physiol. Part D Genom. Proteom. 2006, 1, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Marancik, D.P.; Fast, M.D.; Camus, A.C. Proteomic Characterization of the Acute-Phase Response of Yellow Stingrays Urobatis jamaicensis after Injection with a Vibrio anguillarum-ordalii Bacterin. Fish Shellfish Immunol. 2013, 34, 1383–1389. [Google Scholar] [CrossRef]
- Perry, C.T.; Pratte, Z.A.; Clavere-Graciette, A.; Ritchie, K.B.; Hueter, R.E.; Newton, A.L.; Fischer, G.C.; Dinsdale, E.A.; Doane, M.P.; Wilkinson, K.A.; et al. Elasmobranch Microbiomes: Emerging Patterns and Implications for Host Health and Ecology. Anim. Microbiome 2021, 3, 61. [Google Scholar] [CrossRef]
- Doane, M.P.; Johnson, C.J.; Johri, S.; Kerr, E.N.; Morris, M.M.; Desantiago, R.; Turnlund, A.C.; Goodman, A.; Mora, M.; Lima, L.F.O.; et al. The Epidermal Microbiome Within an Aggregation of Leopard Sharks (Triakis semifasciata) Has Taxonomic Flexibility with Gene Functional Stability Across Three Time-Points. Microb. Ecol. 2022, 1, 3. [Google Scholar] [CrossRef]
- Black, C.; Merly, L.; Hammerschlag, N. Bacterial Communities in Multiple Tissues across the Body Surface of Three Coastal Shark Species. Zool. Stud. 2021, 60, 60–69. [Google Scholar] [CrossRef]
- Stein, R.W.; Mull, C.G.; Kuhn, T.S.; Aschliman, N.C.; Davidson, L.N.K.; Joy, J.B.; Smith, G.J.; Dulvy, N.K.; Mooers, A.O. Global Priorities for Conserving the Evolutionary History of Sharks, Rays and Chimaeras. Nat. Ecol. Evol. 2018, 2, 288–298. [Google Scholar] [CrossRef]
- Marra, N.J.; Stanhope, M.J.; Jue, N.K.; Wang, M.; Sun, Q.; Bitar, P.P.; Richards, V.P.; Komissarov, A.; Rayko, M.; Kliver, S.; et al. White Shark Genome Reveals Ancient Elasmobranch Adaptations Associated with Wound Healing and the Maintenance of Genome Stability. Proc. Natl. Acad. Sci. USA 2019, 116, 4446–4455. [Google Scholar] [CrossRef] [PubMed]
- Marconi, A.; Hancock-Ronemus, A.; Gillis, J.A. Adult Chondrogenesis and Spontaneous Cartilage Repair in the Skate, Leucoraja erinacea. eLife 2020, 9, e53414. [Google Scholar] [CrossRef]
- Alves, L.M.F.; Lemos, M.F.L.; Cabral, H.; Novais, S.C. Elasmobranchs as Bioindicators of Pollution in the Marine Environment. Mar. Pollut. Bull. 2022, 176, 113418. [Google Scholar] [CrossRef] [PubMed]
- Andreev, P.; Coates, M.I.; Karatajute-Talimaa, V.; Shelton, R.M.; Cooper, P.R.; Wang, N.Z.; Sansom, I.J. The Systematics of the Mongolepidida (Chondrichthyes) and the Ordovician Origins of the Clade. PeerJ 2016, 2016, e1850. [Google Scholar] [CrossRef]
- White, W.T.; O’Neill, H.L.; Naylor, G.J.P. Taxonomy and Diversity of Extant Elasmobranchs. In Biology of Sharks and Their Relatives; CRC Press: Boca Raton, FL, USA, 2022; pp. 31–57. ISBN 9781003262190. [Google Scholar]
- Techera, E.J.; Klein, N. Fragmented Governance: Reconciling Legal Strategies for Shark Conservation and Management. Mar. Policy 2011, 35, 73–78. [Google Scholar] [CrossRef]
- Roemer, R.P.; Gallagher, A.J.; Hammerschlag, N. Shallow Water Tidal Flat Use and Associated Specialized Foraging Behavior of the Great Hammerhead Shark (Sphyrna mokarran). Mar. Freshw. Behav. Physiol. 2016, 49, 235–249. [Google Scholar] [CrossRef]
- Weigmann, S. Annotated Checklist of the Living Sharks, Batoids and Chimaeras (Chondrichthyes) of the World, with a Focus on Biogeographical Diversity. J. Fish Biol. 2016, 88, 837–1037. [Google Scholar] [CrossRef]
- Engelbrecht, A.; Mörs, T.; Reguero, M.A.; Kriwet, J. Skates and Rays (Elasmobranchii, Batomorphii) from the Eocene La Meseta and Submeseta Formations, Seymour Island, Antarctica. Hist. Biol. 2018, 31, 1028–1044. [Google Scholar] [CrossRef]
- Pikitch, E.K.; Chapman, D.D.; Babcock, E.A.; Shivji, M.S. Habitat Use and Demographic Population Structure of Elasmobranchs at a Caribbean Atoll (Glover’s Reef, Belize). Mar. Ecol. Prog. Ser. 2005, 302, 187–197. [Google Scholar] [CrossRef]
- Ruocco, N.L.; Lucifora, L.O. Ecological Singularity of Temperate Mesopredatory Myliobatoid Rays (Chondrichthyes: Myliobatiformes). Mar. Freshw. Res. 2017, 68, 1098. [Google Scholar] [CrossRef]
- Myers, R.A.; Baum, J.K.; Shepherd, T.D.; Powers, S.P.; Peterson, C.H. Cascading Effects of the Loss of Apex Predatory Sharks from a Coastal Ocean. Science 2007, 315, 1846–1850. [Google Scholar] [CrossRef] [PubMed]
- Moss, S. Feeding Mechanisms in Sharks. Integr. Comp. Biol. 1977, 17, 355–364. [Google Scholar] [CrossRef]
- Misty Paig-Tran, E.W.; Summers, A.P. Comparison of the Structure and Composition of the Branchial Filters in Suspension Feeding Elasmobranchs. Anat. Rec. 2014, 297, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Dulvy, N.K.; Pacoureau, N.; Rigby, C.L.; Pollom, R.A.; Jabado, R.W.; Ebert, D.A.; Finucci, B.; Pollock, C.M.; Cheok, J.; Derrick, D.H.; et al. Overfishing Drives over One-Third of All Sharks and Rays toward a Global Extinction Crisis. Curr. Biol. 2021, 31, 4773–4787.e8. [Google Scholar] [CrossRef] [PubMed]
- DiBattista, J.D. Patterns of Genetic Variation in Anthropogenically Impacted Populations. Conserv. Genet. 2008, 9, 141–156. [Google Scholar] [CrossRef]
- Liu, K.M.; Wu, C.B.; Joung, S.J.; Tsai, W.P.; Su, K.Y. Multi-Model Approach on Growth Estimation and Association With Life History Trait for Elasmobranchs. Front. Mar. Sci. 2021, 8, 108. [Google Scholar] [CrossRef]
- Nielsen, J.; Hedeholm, R.B.; Heinemeier, J.; Bushnell, P.G.; Christiansen, J.S.; Olsen, J.; Ramsey, C.B.; Brill, R.W.; Simon, M.; Steffensen, K.F.; et al. Eye Lens Radiocarbon Reveals Centuries of Longevity in the Greenland Shark (Somniosus microcephalus). Science 2016, 353, 702–704. [Google Scholar] [CrossRef]
- Dent, F.; Clarke, S. State of the Global Market for Shark Products; FAO Fishereis and Aquaculture Technical Paper, No. 590; FAO: Rome, Italy, 2015; p. 187. [Google Scholar]
- Bornatowski, H.; Braga, R.R.; Vitule, J.R.S.; Simões Vitule, J.R. Shark Mislabeling Threatens Biodiversity. Science 2013, 340, 923. [Google Scholar] [CrossRef]
- Pazartzi, T.; Siaperopoulou, S.; Gubili, C.; Maradidou, S.; Loukovitis, D.; Chatzispyrou, A.; Griffiths, A.M.; Minos, G.; Imsiridou, A. High Levels of Mislabeling in Shark Meat—Investigating Patterns of Species Utilization with DNA Barcoding in Greek Retailers. Food Control 2019, 98, 179–186. [Google Scholar] [CrossRef]
- Lack, M.; Sant, G. Illegal, Unreported and Unregulated Shark Catch: A Review of Current Knowledge and Action; Traffic: Canaberra, Australia, 2008; p. 62. [Google Scholar]
- Mucientes, G.; Vedor, M.; Sims, D.W.; Queiroz, N. Unreported Discards of Internationally Protected Pelagic Sharks in a Global Fishing Hotspot Are Potentially Large. Biol. Conserv. 2022, 269, 109534. [Google Scholar] [CrossRef]
- Baeta, F.; Batista, M.; Maia, A.; Costa, M.J.; Cabral, H. Elasmobranch Bycatch in a Trammel Net Fishery in the Portuguese West Coast. Fish. Res. 2010, 102, 123–129. [Google Scholar] [CrossRef]
- Storai, T.; Zinzula, L.; Repetto, S.; Zuffa, M.; Morgan, A.; Mandelman, J. Bycatch of Large Elasmobranchs in the Traditional Tuna Traps (Tonnare) of Sardinia from 1990 to 2009. Fish. Res. 2011, 109, 74–79. [Google Scholar] [CrossRef]
- Cross, H. Elasmobranch Capture by Commercial Small-Scale Fisheries in the Bijagós Archipelago, Guinea Bissau. Fish. Res. 2015, 168, 105–108. [Google Scholar] [CrossRef]
- Spaet, J.L.Y.; Berumen, M.L. Fish Market Surveys Indicate Unsustainable Elasmobranch Fisheries in the Saudi Arabian Red Sea. Fish. Res. 2015, 161, 356–364. [Google Scholar] [CrossRef]
- Navia, A.F.; Mejía-Falla, P.A. Fishing Effects on Elasmobranchs from the Pacific Coast of Colombia. Univ. Sci. 2016, 21, 9–22. [Google Scholar] [CrossRef]
- Lynch, A.M.J.; Sutton, S.G.; Simpfendorfer, C.A. Implications of Recreational Fishing for Elasmobranch Conservation in the Great Barrier Reef Marine Park. Aquat. Conserv. 2010, 20, 312–318. [Google Scholar] [CrossRef]
- Gallagher, A.J.; Hammerschlag, N.; Danylchuk, A.J.; Cooke, S.J. Shark Recreational Fisheries: Status, Challenges, and Research Needs. Ambio 2017, 46, 385–398. [Google Scholar] [CrossRef]
- Cardeñosa, D. Genetic Identification of Threatened Shark Species in Pet Food and Beauty Care Products. Conserv. Genet. 2019, 20, 1383–1387. [Google Scholar] [CrossRef]
- Diogo, G.S.; Carneiro, F.; Freitas-Ribeiro, S.; Sotelo, C.G.; Pérez-Martín, R.I.; Pirraco, R.P.; Reis, R.L.; Silva, T.H. Prionace Glauca Skin Collagen Bioengineered Constructs as a Promising Approach to Trigger Cartilage Regeneration. Mater. Sci. Eng. C 2020, 120, 111587. [Google Scholar] [CrossRef]
- Seixas, M.J.; Martins, E.; Reis, R.L.; Silva, T.H. Extraction and Characterization of Collagen from Elasmobranch Byproducts for Potential Biomaterial Use. Mar. Drugs 2020, 18, 617. [Google Scholar] [CrossRef]
- Cho, J.J.; Kim, Y.T. Sharks: A Potential Source of Antiangiogenic Factors and Tumor Treatments. Mar. Biotechnol. 2002, 4, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Rabbani-chadegani, A.; Abdossamadi, S.; Bargahi, A.; Yousef-masboogh, M. Identification of Low-Molecular-Weight Protein (SCP1) from Shark Cartilage with Anti-Angiogenesis Activity and Sequence Similarity to Parvalbumin. J. Pharm. Biomed. Anal. 2008, 46, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Ostrander, G.K.; Cheng, K.C.; Wolf, J.C.; Wolfe, M.J. Shark Cartilage, Cancer and the Growing Threat of Pseudoscience. Cancer Res. 2004, 64, 8485–8491. [Google Scholar] [CrossRef] [PubMed]
- Sansom, I.J.; Davies, N.S.; Coates, M.I.; Nicoll, R.S.; Ritchie, A. Chondrichthyan-like Scales from the Middle Ordovician of Australia. Palaeontology 2012, 55, 243–247. [Google Scholar] [CrossRef]
- Whitenack, L.B.; Kim, S.L.; Sibert, E.C. Bridging the Gap Between Chondrichthyan Paleobiology and Biology. In Biology of Sharks and Their Relatives; CRC Press: Boca Raton, FL, USA, 2022; pp. 1–29. [Google Scholar] [CrossRef]
- King, B.L.; Gillis, J.A.; Carlisle, H.R.; Dahn, R.D. A Natural Deletion of the HoxC Cluster in Elasmobranch Fishes. Science 2011, 334, 1517. [Google Scholar] [CrossRef]
- Domingues, R.R.; Hilsdorf, A.W.S.; Gadig, O.B.F. The Importance of Considering Genetic Diversity in Shark and Ray Conservation Policies. Conserv. Genet. 2018, 19, 501–525. [Google Scholar] [CrossRef]
- Wang, B.; Tseng, E.; Regulski, M.; Clark, T.A.; Hon, T.; Jiao, Y.; Lu, Z.; Olson, A.; Stein, J.C.; Ware, D. Unveiling the Complexity of the Maize Transcriptome by Single-Molecule Long-Read Sequencing. Nat. Commun. 2016, 7, 11708. [Google Scholar] [CrossRef]
- Tilgner, H.; Grubert, F.; Sharon, D.; Snyder, M.P. Defining a Personal, Allele-Specific, and Single-Molecule Long-Read Transcriptome. Proc. Natl. Acad. Sci. USA 2014, 111, 9869–9874. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; Weissensteiner, M.H.; Sedlazeck, F.J. Towards Population-Scale Long-Read Sequencing. Nat. Rev. Genet. 2021, 22, 572–587. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. The Many Pathways of RNA Degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef]
- RNALater TM Tissue Collection: RNA Stabilization Solution. In Ambion Protocols and Manuals; Life Technologies Corporation: Carlsbad, CA, USA, 2011.
- Fabre, A.L.; Colotte, M.; Luis, A.; Tuffet, S.; Bonnet, J. An Efficient Method for Long-Term Room Temperature Storage of RNA. Eur. J. Hum. Genet. 2014, 22, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. The Review of Transcriptome Sequencing: Principles, History and Advances. IOP Conf. Ser. Earth Environ. Sci. 2019, 332, 042003. [Google Scholar] [CrossRef]
- Schadt, E.E.; Turner, S.; Kasarskis, A. A Window into Third-Generation Sequencing. Hum. Mol. Genet. 2010, 19, R227–R240. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.; Prenzler, A.; Eils, R.; von der Schulenburg, J.M.G. Genome Sequencing: A Systematic Review of Health Economic Evidence. Health Econ. Rev. 2013, 3, 29. [Google Scholar] [CrossRef] [PubMed]
- von Bubnoff, A. Next-Generation Sequencing: The Race Is On. Cell 2008, 132, 721–723. [Google Scholar] [CrossRef]
- McCombie, W.R.; McPherson, J.D.; Mardis, E.R. Next-Generation Sequencing Technologies. Cold Spring Harb. Perspect. Med. 2019, 9, a036798. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Ba, Y.; Zhuang, Q.; Zhong, G. RNA-Seq Technology and Its Application in Fish Transcriptomics. OMICS 2014, 18, 98–110. [Google Scholar] [CrossRef]
- Nagalakshmi, U.; Waern, K.; Snyder, M. RNA-Seq: A Method for Comprehensive Transcriptome Analysis. Curr. Protoc. Mol. Biol. 2010, 89, 4.11.1–4.11.13. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of Next-Generation Sequencing Systems. J. Biomed. Biotechnol. 2012, 2012, 251364. [Google Scholar] [CrossRef]
- Husseini, A.A.; Derakhshandeh, M.; Tatlisu, N.B. Comprehensive Review of Transcriptomics (RNAs) Workflows from Blood Specimens. Sep. Purif. Rev. 2022, 51, 57–77. [Google Scholar] [CrossRef]
- Akbar, M.A.; Ahmad, A.; Usup, G.; Bunawan, H. RNA-Seq as an Emerging Tool for Marine Dinoflagellate Transcriptome Analysis: Process and Challenges. Processes 2018, 6, 5. [Google Scholar] [CrossRef]
- Malone, J.H.; Oliver, B. Microarrays, Deep Sequencing and the True Measure of the Transcriptome. BMC Biol. 2011, 9, 34. [Google Scholar] [CrossRef]
- Rodríguez-Jorquera, I.A.; Colli-Dula, R.C.; Kroll, K.; Jayasinghe, B.S.; Parachu Marco, M.V.; Silva-Sanchez, C.; Toor, G.S.; Denslow, N.D. Blood Transcriptomics Analysis of Fish Exposed to Perfluoro Alkyls Substances: Assessment of a Non-Lethal Sampling Technique for Advancing Aquatic Toxicology Research. Environ. Sci. Technol. 2019, 53, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Arteaga, A.; Wu, Y.; Silva-Marrero, J.I.; Rashidpour, A.; Almajano, M.P.; Fernández, F.; Baanante, I.V.; Metón, I. Gene Markers of Dietary Macronutrient Composition and Growth in the Skeletal Muscle of Gilthead Sea Bream (Sparus aurata). Aquaculture 2022, 555, 738221. [Google Scholar] [CrossRef]
- Martin, S.A.M.; Dehler, C.E.; Król, E. Transcriptomic Responses in the Fish Intestine. Dev. Comp. Immunol. 2016, 64, 103–117. [Google Scholar] [CrossRef]
- Jaksik, R.; Iwanaszko, M.; Rzeszowska-Wolny, J.; Kimmel, M. Microarray Experiments and Factors Which Affect Their Reliability. Biol. Direct 2015, 10, 46. [Google Scholar] [CrossRef]
- Molina-Olvera, G.; Rivas-Ortiz, C.I.; Schcolnik-Cabrera, A.; Castillo-Rodal, A.I.; López-Vidal, Y. RNA Microarray-Based Comparison of Innate Immune Phenotypes between Human THP-1 Macrophages Stimulated with Two BCG Strains. Int. J. Mol. Sci. 2022, 23, 4525. [Google Scholar] [CrossRef]
- Zhao, S.; Xu, J.; Zhang, W.; Yan, W.; Li, G. Paternal Exposure to Microcystin-LR Triggers Developmental Neurotoxicity in Zebrafish Offspring via an Epigenetic Mechanism Involving MAPK Pathway. Sci. Total Environ. 2021, 792, 148437. [Google Scholar] [CrossRef]
- Bracamonte, A.G. Microarrays towards Nanoarrays and the Future Next Generation of Sequencing Methodologies (NGS). Sens. Biosensing Res. 2022, 37, 100503. [Google Scholar] [CrossRef]
- Bleidorn, C. Third Generation Sequencing: Technology and Its Potential Impact on Evolutionary Biodiversity Research. Syst. Biodivers. 2016, 14, 1–8. [Google Scholar] [CrossRef]
- Smith, M.A.; Ersavas, T.; Ferguson, J.M.; Liu, H.; Lucas, M.C.; Begik, O.; Bojarski, L.; Barton, K.; Novoa, E.M. Barcoding and Demultiplexing Oxford Nanopore Native RNA Sequencing Reads with Deep Residual Learning. bioRxiv 2019, 864322. [Google Scholar] [CrossRef]
- Cook, D.E.; Valle-Inclan, J.E.; Pajoro, A.; Rovenich, H.; Thomma, B.P.H.J.; Faino, L. Long-Read Annotation: Automated Eukaryotic Genome Annotation Based on Long-Read CDNA Sequencing. Plant Physiol. 2019, 179, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Winefield, C.; Bombarely, A.; Prentis, P.; Waterhouse, P. Tools and Strategies for Long-Read Sequencing and De Novo Assembly of Plant Genomes. Trends Plant Sci. 2019, 24, 700–724. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Tang, F. Recent Advances in Single-Cell Sequencing Technologies. Precis. Clin. Med. 2022, 5, 700–724. [Google Scholar] [CrossRef] [PubMed]
- Rang, F.J.; Kloosterman, W.P.; de Ridder, J. From Squiggle to Basepair: Computational Approaches for Improving Nanopore Sequencing Read Accuracy. Genome Biol. 2018, 19, 90. [Google Scholar] [CrossRef]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-Generation Sequencing Technologies: An Overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate Circular Consensus Long-Read Sequencing Improves Variant Detection and Assembly of a Human Genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Hara, Y.; Yamaguchi, K.; Onimaru, K.; Kadota, M.; Koyanagi, M.; Keeley, S.D.; Tatsumi, K.; Tanaka, K.; Motone, F.; Kageyama, Y.; et al. Shark Genomes Provide Insights into Elasmobranch Evolution and the Origin of Vertebrates. Nat. Ecol. Evol. 2018, 2, 1761–1771. [Google Scholar] [CrossRef]
- Swenson, J.D.; Klomp, J.; Fisher, R.A.; Crow, K.D. How the Devil Ray Got Its Horns: The Evolution and Development of Cephalic Lobes in Myliobatid Stingrays (Batoidea: Myliobatidae). Front. Ecol. Evol. 2018, 6, 181. [Google Scholar] [CrossRef]
- Mayeur, H.; Lanoizelet, M.; Quillien, A.; Menuet, A.; Michel, L.; Martin, K.J.; Dejean, S.; Blader, P.; Mazan, S.; Lagadec, R. When Bigger Is Better: 3D RNA Profiling of the Developing Head in the Catshark Scyliorhinus canicula. Front. Cell Dev. Biol. 2021, 9, 2944. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, K.M.; Teffer, A.; Michaleski, S.; Bernier, N.J.; Heath, D.D.; Miller, K.M. The Use of Non-Lethal Sampling for Transcriptomics to Assess the Physiological Status of Wild Fishes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2021, 256, 110629. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, B.; Kirkness, E.F.; Loh, Y.H.; Halpern, A.L.; Lee, A.P.; Johnson, J.; Dandona, N.; Viswanathan, L.D.; Tay, A.; Venter, J.C.; et al. Survey Sequencing and Comparative Analysis of the Elephant Shark (Callorhinchus milii) Genome. PLoS Biol. 2007, 5, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Grunow, B.; Reismann, T.; Moritz, T. Pre-Hatching Ontogenetic Changes of Morphological Characters of Small-Spotted Catshark (Scyliorhinus canicula). Fishes 2022, 7, 100. [Google Scholar] [CrossRef]
- Harahush, B.K.; Fischer, A.B.P.; Collin, S.P. Captive Breeding and Embryonic Development of Chiloscyllium punctatum Muller & Henle, 1838 (Elasmobranchii: Hemiscyllidae). J. Fish Biol. 2007, 71, 1007–1022. [Google Scholar] [CrossRef]
- Robinson, D.P.; Baverstock, W.; Al-Jaru, A.; Hyland, K.; Khazanehdari, K.A. Annually Recurring Parthenogenesis in a Zebra Shark Stegostoma Fasciatum. J. Fish Biol. 2011, 79, 1376–1382. [Google Scholar] [CrossRef]
- Honda, Y.; Takagi, W.; Wong, M.K.S.; Ogawa, N.; Tokunaga, K.; Kofuji, K.; Hyodo, S. Morphological and Functional Development of the Spiral Intestine in Cloudy Catshark (Scyliorhinus torazame). J. Exp. Biol. 2020, 223, jeb225557. [Google Scholar] [CrossRef]
- Boisvert, C.A.; Martins, C.L.; Edmunds, A.G.; Cocks, J.; Currie, P. Capture, Transport, and Husbandry of Elephant Sharks (Callorhinchus milii) Adults, Eggs, and Hatchlings for Research and Display. Zoo Biol. 2014, 34, 94–98. [Google Scholar] [CrossRef]
- Saha, S.; Sparks, A.B.; Rago, C.; Akmaev, V.; Wang, C.J.; Vogelstein, B.; Kinzler, K.W.; Velculescu, V.E. Using the Transcriptome to Annotate the Genome. Nat. Biotechnol. 2002, 20, 508–512. [Google Scholar] [CrossRef]
- Lou, F.; Wang, L.; Wang, Z.; Wang, L.; Zhao, L.; Zhou, Q.; Lu, Z.; Tang, Y. Full-Length Transcriptome of the Whale Shark (Rhincodon typus) Facilitates the Genome Information. Front. Mar. Sci. 2022, 8, 2089. [Google Scholar] [CrossRef]
- Shields, E.J.; Sorida, M.; Sheng, L.; Sieriebriennikov, B.; Ding, L.; Bonasio, R. Genome Annotation with Long RNA Reads Reveals New Patterns of Gene Expression and Improves Single-Cell Analyses in an Ant Brain. BMC Biol. 2021, 19, 254. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Collins, R.L.; Lee, W.P.; Weber, A.M.; Jun, Y.; Zhu, Q.; Weisburd, B.; Huang, Y.; Audano, P.A.; Wang, H.; et al. Expectations and Blind Spots for Structural Variation Detection from Long-Read Assemblies and Short-Read Genome Sequencing Technologies. Am. J. Hum. Genet. 2021, 108, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Suleski, M.; Craig, J.M.; Kasprowicz, A.E.; Sanderford, M.; Li, M.; Stecher, G.; Hedges, S.B. TimeTree 5: An Expanded Resource for Species Divergence Times. Mol. Biol. Evol. 2022, 39, msac174. [Google Scholar] [CrossRef] [PubMed]
- Takechi, M.; Takeuchi, M.; Ota, K.G.; Nishimura, O.; Mochii, M.; Itomi, K.; Adachi, N.; Takahashi, M.; Fujimoto, S.; Tarui, H.; et al. Overview of the Transcriptome Profiles Identified in Hagfish, Shark, and Bichir: Current Issues Arising from Some Nonmodel Vertebrate Taxa. J. Exp. Zool. B Mol. Dev. Evol. 2011, 316B, 526–546. [Google Scholar] [CrossRef]
- Richards, V.P.; Suzuki, H.; Stanhope, M.J.; Shivji, M.S. Characterization of the Heart Transcriptome of the White Shark (Carcharodon carcharias). BMC Genom. 2013, 14, 697. [Google Scholar] [CrossRef]
- Youson, J.H.; Al-Mahrouki, A.A. Ontogenetic and Phylogenetic Development of the Endocrine Pancreas (Islet Organ) in Fishes. Gen. Comp. Endocrinol. 1999, 116, 303–335. [Google Scholar] [CrossRef]
- Mulley, J.F.; Hargreaves, A.D.; Hegarty, M.J.; Heller, R.S.; Swain, M.T. Transcriptomic Analysis of the Lesser Spotted Catshark (Scyliorhinus canicula) Pancreas, Liver and Brain Reveals Molecular Level Conservation of Vertebrate Pancreas Function. BMC Genom. 2014, 15, 1074. [Google Scholar] [CrossRef]
- Arntfield, M.E.; van der Kooy, D. β-Cell Evolution: How the Pancreas Borrowed from the Brain: The Shared Toolbox of Genes Expressed by Neural and Pancreatic Endocrine Cells May Reflect Their Evolutionary Relationship. BioEssays 2011, 33, 582–587. [Google Scholar] [CrossRef]
- Yamamoto, K.; Bloch, S.; Vernier, P. New Perspective on the Regionalization of the Anterior Forebrain in Osteichthyes. Dev. Growth Differ. 2017, 59, 175–187. [Google Scholar] [CrossRef]
- Lamb, T.D.; Patel, H.; Chuah, A.; Natoli, R.C.; Davies, W.I.L.; Hart, N.S.; Collin, S.P.; Hunt, D.M. Evolution of Vertebrate Phototransduction: Cascade Activation. Mol. Biol. Evol. 2016, 33, 2064–2087. [Google Scholar] [CrossRef]
- Domingues, R.R.; Mastrochirico-Filho, V.A.; Mendes, N.J.; Hashimoto, D.T.; Coelho, R.; da Cruz, V.P.; Antunes, A.; Foresti, F.; Mendonça, F.F. Comparative Eye and Liver Differentially Expressed Genes Reveal Monochromatic Vision and Cancer Resistance in the Shortfin Mako Shark (Isurus oxyrinchus). Genomics 2020, 112, 4817–4826. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Núñez, I.; Robledo, D.; Mayeur, H.; Mazan, S.; Sánchez, L.; Adrio, F.; Barreiro-Iglesias, A.; Candal, E. Loss of Active Neurogenesis in the Adult Shark Retina. Front. Cell Dev. Biol. 2021, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Marra, N.J.; Richards, V.P.; Early, A.; Bogdanowicz, S.M.; Pavinski Bitar, P.D.; Stanhope, M.J.; Shivji, M.S. Comparative Transcriptomics of Elasmobranchs and Teleosts Highlight Important Processes in Adaptive Immunity and Regional Endothermy. BMC Genom. 2017, 18, 87. [Google Scholar] [CrossRef] [PubMed]
- Swift, D.G.; Dunning, L.T.; Igea, J.; Brooks, E.J.; Jones, C.S.; Noble, L.R.; Ciezarek, A.; Humble, E.; Savolainen, V. Evidence of Positive Selection Associated with Placental Loss in Tiger Sharks. BMC Evol. Biol. 2016, 16, 126. [Google Scholar] [CrossRef]
- Opazo, J.C.; Lee, A.P.; Hoffmann, F.G.; Toloza-Villalobos, J.; Burmester, T.; Venkatesh, B.; Storz, J.F. Ancient Duplications and Expression Divergence in the Globin Gene Superfamily of Vertebrates: Insights from the Elephant Shark Genome and Transcriptome. Mol. Biol. Evol. 2015, 32, 1684–1694. [Google Scholar] [CrossRef]
- Ciezarek, A.G.; Dunning, L.T.; Jones, C.S.; Noble, L.R.; Humble, E.; Stefanni, S.S.; Savolainen, V. Substitutions in the Glycogenin-1 Gene Are Associated with the Evolution of Endothermy in Sharks and Tunas. Genome Biol. Evol. 2016, 8, 3011–3021. [Google Scholar] [CrossRef]
- Goshima, M.; Sekiguchi, R.; Matsushita, M.; Nonaka, M. The Complement System of Elasmobranches Revealed by Liver Transcriptome Analysis of a Hammerhead Shark, Sphyrna zygaena. Dev. Comp. Immunol. 2016, 61, 13–24. [Google Scholar] [CrossRef]
- Martinez, C.M.; Rohlf, F.J.; Frisk, M.G. Re-Evaluation of Batoid Pectoral Morphology Reveals Novel Patterns of Diversity among Major Lineages. J. Morphol. 2016, 277, 482–493. [Google Scholar] [CrossRef]
- Nakamura, T.; Klomp, J.; Pieretti, J.; Schneider, I.; Gehrke, A.R.; Shubin, A.N.H. Molecular Mechanisms Underlying the Exceptional Adaptations of Batoid Fins. Proc. Natl. Acad. Sci. USA 2015, 112, 15940–15945. [Google Scholar] [CrossRef]
- Hall, K.C.; Hundt, P.J.; Swenson, J.D.; Summers, A.P.; Crow, K.D. The Evolution of Underwater Flight: The Redistribution of Pectoral Fin Rays, in Manta Rays and Their Relatives (Myliobatidae). J. Morphol. 2018, 279, 1155–1170. [Google Scholar] [CrossRef]
- Junker, J.P.; Noël, E.S.; Guryev, V.; Peterson, K.A.; Shah, G.; Huisken, J.; McMahon, A.P.; Berezikov, E.; Bakkers, J.; Van Oudenaarden, A. Genome-Wide RNA Tomography in the Zebrafish Embryo. Cell 2014, 159, 662–675. [Google Scholar] [CrossRef] [PubMed]
- Combs, P.A.; Eisen, M.B. Genome-Wide Measurement of Spatial Expression in Patterning Mutants of Drosophila Melanogaster. F1000Research 2017, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Baek, M.; D’Elia, K.P.; Boisvert, C.; Currie, P.D.; Tay, B.H.; Venkatesh, B.; Brown, S.M.; Heguy, A.; Schoppik, D.; et al. The Ancient Origins of Neural Substrates for Land Walking. Cell 2018, 172, 667–682.e15. [Google Scholar] [CrossRef]
- Onimaru, K.; Tatsumi, K.; Tanegashima, C.; Kadota, M.; Nishimura, O.; Kuraku, S. Developmental Hourglass and Heterochronic Shifts in Fin and Limb Development. eLife 2021, 10, e62865. [Google Scholar] [CrossRef] [PubMed]
- Honda, Y.; Ogawa, N.; Wong, M.K.S.; Tokunaga, K.; Kuraku, S.; Hyodo, S.; Takagi, W. Molecular Mechanism of Nutrient Uptake in Developing Embryos of Oviparous Cloudy Catshark (Scyliorhinus torazame). PLoS ONE 2022, 17, e0265428. [Google Scholar] [CrossRef] [PubMed]
- Wood, C.M.; Walsh, P.J.; Kajimura, M.; McClelland, G.B.; Chew, S.F. The Influence of Feeding and Fasting on Plasma Metabolites in the Dogfish Shark (Squalus acanthias). Comp. Biochem. Physiol.-Mol. Integr. Physiol. 2010, 155, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Deck, C.A.; McKay, S.J.; Fiedler, T.J.; Lemoine, C.M.R.; Kajimura, M.; Nawata, C.M.; Wood, C.M.; Walsh, P.J. Transcriptome Responses in the Rectal Gland of Fed and Fasted Spiny Dogfish Shark (Squalus acanthias) Determined by Suppression Subtractive Hybridization. Comp. Biochem. Physiol. Part D Genom. Proteom. 2013, 8, 334–343. [Google Scholar] [CrossRef]
- Chana-Munoz, A.; Jendroszek, A.; Sønnichsen, M.; Kristiansen, R.; Jensen, J.K.; Andreasen, P.A.; Bendixen, C.; Panitz, F. Multi-Tissue RNA-Seq and Transcriptome Characterisation of the Spiny Dogfish Shark (Squalus acanthias) Provides a Molecular Tool for Biological Research and Reveals New Genes Involved in Osmoregulation. PLoS ONE 2017, 12, e0182756. [Google Scholar] [CrossRef]
- Matthews, G.D.; Gould, R.M.; Vardimon, L. A Single Glutamine Synthetase Gene Produces Tissue-Specific Subcellular Localization by Alternative Splicing. FEBS Lett. 2005, 579, 5527–5534. [Google Scholar] [CrossRef]
- Wu, G.; Morris, S.M. Arginine Metabolism: Nitric Oxide and Beyond. Biochem. J. 1998, 336, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Knepper, M.A.; Wade, J.B.; Terris, J.; Ecelbarger, C.A.; Marples, D.; Mandon, B.; Chou, C.L.; Kishore, B.K.; Nielsen, S. Renal Aquaporins. Kidney Int. 1996, 49, 1712–1717. [Google Scholar] [CrossRef] [PubMed]
- Pillans, R.D.; Good, J.P.; Gary Anderson, W.; Hazon, N.; Franklin, C.E. Freshwater to Seawater Acclimation of Juvenile Bull Sharks (Carcharhinus leucas): Plasma Osmolytes and Na+/K+-ATPase Activity in Gill, Rectal Gland, Kidney and Intestine. J. Comp. Physiol. B 2005, 175, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Imaseki, I.; Wakabayashi, M.; Hara, Y.; Watanabe, T.; Takabe, S.; Kakumura, K.; Honda, Y.; Ueda, K.; Murakumo, K.; Matsumoto, R.; et al. Comprehensive Analysis of Genes Contributing to Euryhalinity in the Bull Shark, Carcharhinus leucas; Na+-Cl− Co-Transporter Is One of the Key Renal Factors up-Regulated in Acclimation to Low-Salinity Environment in Bull Sharks, but Not in Houndsharks, Tr. J. Exp. Biol. 2019, 222, jeb.201780. [Google Scholar] [CrossRef] [PubMed]
- Fenton, R.A.; Knepper, M.A. Mouse Models and the Urinary Concentrating Mechanism in the New Millennium. Physiol. Rev. 2007, 87, 1083–1112. [Google Scholar] [CrossRef] [PubMed]
- Aburatani, N.; Takagi, W.; Wong, M.K.S.; Kadota, M.; Kuraku, S.; Tokunaga, K.; Kofuji, K.; Saito, K.; Godo, W.; Sakamoto, T.; et al. Facilitated NaCl Uptake in the Highly Developed Bundle of the Nephron in Japanese Red Stingray Hemitrygon Akajei Revealed by Comparative Anatomy and Molecular Mapping. Zool. Sci. 2020, 37, 458–466. [Google Scholar] [CrossRef]
- Lighten, J.; Incarnato, D.; Ward, B.J.; van Oosterhout, C.; Bradbury, I.; Hanson, M.; Bentzen, P. Adaptive Phenotypic Response to Climate Enabled by Epigenetics in a K-Strategy Species, the Fish Leucoraja Ocellata (Rajidae). R. Soc. Open Sci. 2016, 3, 160299. [Google Scholar] [CrossRef]
- Petitjean, Q.; Jean, S.; Gandar, A.; Côte, J.; Laffaille, P.; Jacquin, L. Stress Responses in Fish: From Molecular to Evolutionary Processes. Sci. Total Environ. 2019, 684, 371–380. [Google Scholar] [CrossRef]
- Nazarian, J.; Hathout, Y.; Vertes, A.; Hoffman, E.P. The Proteome Survey of an Electricity-Generating Organ (Torpedo californica Electric Organ). Proteomics 2007, 7, 617–627. [Google Scholar] [CrossRef]
- Stavrianakou, M.; Perez, R.; Wu, C.; Sachs, M.S.; Aramayo, R.; Harlow, M. Draft de Novo Transcriptome Assembly and Proteome Characterization of the Electric Lobe of Tetronarce Californica: A Molecular Tool for the Study of Cholinergic Neurotransmission in the Electric Organ. BMC Genom. 2017, 18, 611. [Google Scholar] [CrossRef]
- Newton, K.C.; Gill, A.B.; Kajiura, S.M. Electroreception in Marine Fishes: Chondrichthyans. J. Fish Biol. 2019, 95, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Bellono, N.W.; Leitch, D.B.; Julius, D. Molecular Basis of Ancestral Vertebrate Electroreception. Nature 2017, 543, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Claes, J.M.; Partridge, J.C.; Hart, N.S.; Garza-Gisholt, E.; Ho, H.C.; Mallefet, J.; Collin, S.P. Photon Hunting in the Twilight Zone: Visual Features of Mesopelagic Bioluminescent Sharks. PLoS ONE 2014, 9, e104213. [Google Scholar] [CrossRef] [PubMed]
- Delroisse, J.; Duchatelet, L.; Flammang, P.; Mallefet, J. De Novo Transcriptome Analyses Provide Insights into Opsin-Based Photoreception in the Lanternshark Etmopterus Spinax. PLoS ONE 2018, 13, e0209767. [Google Scholar] [CrossRef] [PubMed]
- Claes, J.M.; Mallefet, J.; Mallefet, J. Bioluminescence of Sharks: First Synthesis. In Bioluminescence in Focus—A Collection of Illuminating Essays; Research Signpost: Kerala, India, 2009; pp. 51–65. [Google Scholar]
- Viviani, V.R. The Origin, Diversity, and Structure Function Relationships of Insect Luciferases. Cell. Mol. Life Sci. 2002, 59, 1833–1850. [Google Scholar] [CrossRef]
- Delroisse, J.; Duchatelet, L.; Flammang, P.; Mallefet, J. Photophore Distribution and Enzymatic Diversity Within the Photogenic Integument of the Cookie-Cutter Shark Isistius brasiliensis (Chondrichthyes: Dalatiidae). Front. Mar. Sci. 2021, 8, 366. [Google Scholar] [CrossRef]
- Riggs, C.L.; Summers, A.; Warren, D.E.; Nilsson, G.E.; Lefevre, S.; Dowd, W.W.; Milton, S.; Podrabsky, J.E. Small Non-Coding RNA Expression and Vertebrate Anoxia Tolerance. Front. Genet. 2018, 9, 230. [Google Scholar] [CrossRef]
- Walker, A.A.; Robinson, S.D.; Hamilton, B.F.; Undheim, E.A.B.; King, G.F. Deadly Proteomes: A Practical Guide to Proteotranscriptomics of Animal Venoms. Proteomics 2020, 20, 1900324. [Google Scholar] [CrossRef]
- Lewis, R.J.; Garcia, M.L. Therapeutic Potential of Venom Peptides. Nat. Rev. Drug Discov. 2003, 2, 790–802. [Google Scholar] [CrossRef]
- Júnior, N.G.D.O.; Fernandes, G.D.R.; Cardoso, M.H.; Costa, F.F.; Cândido, E.D.S.; Neto, D.G.; Mortari, M.R.; Schwartz, E.F.; Franco, O.L.; De Alencar, S.A. Venom Gland Transcriptome Analyses of Two Freshwater Stingrays (Myliobatiformes: Potamotrygonidae) from Brazil. Sci. Rep. 2016, 6, 21935. [Google Scholar] [CrossRef]
- Silva, F.; Huang, Y.; Yang, V.; Mu, X.; Shi, Q.; Antunes, A. Transcriptomic Characterization of the South American Freshwater Stingray Potamotrygon Motoro Venom Apparatus. Toxins 2018, 10, 544. [Google Scholar] [CrossRef] [PubMed]
- Castro, H.C.; Zingali, R.B.; Albuquerque, M.G.; Pujol-Luz, M.; Rodrigues, C.R. Snake Venom Thrombin-like Enzymes: From Reptilase to Now. Cell. Mol. Life Sci. 2004, 61, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Kemparaju, K.; Girish, K.S. Snake Venom Hyaluronidase: A Therapeutic Target. Cell. Biochem. Funct. 2006, 24, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Lomonte, B. Phospholipases A2: Unveiling the Secrets of a Functionally Versatile Group of Snake Venom Toxins. Toxicon 2013, 62, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M.T. Timeline of Key Events in Snake Venom Metalloproteinase Research. J. Proteom. 2009, 72, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, Y.; Morita, T. Structure and Function of Snake Venom Cysteine-Rich Secretory Proteins. Toxicon 2004, 44, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Johnson, W.E.; O’Brien, S.J.; Vasconcelos, V.; Antunes, A. Evolution of CRISPs Associated with Toxicoferan-Reptilian Venom and Mammalian Reproduction. Mol. Biol. Evol. 2012, 29, 1807–1822. [Google Scholar] [CrossRef] [PubMed]
- Yuh, F.P.; Wong, P.T.H.; Kumar, P.P.; Hodgson, W.C.; Kini, R.M. Ohanin, a Novel Protein from King Cobra Venom, Induces Hypolocomotion and Hyperalgesia in Mice. J. Biol. Chem. 2005, 280, 13137–13147. [Google Scholar] [CrossRef]
- Dunbar, J.P.; Fort, A.; Redureau, D.; Sulpice, R.; Dugon, M.M.; Quinton, L. Venomics Approach Reveals a High Proportion of Lactrodectus-like Toxins in the Venom of the Noble Falsewidow Spider Steatoda Nobilis. Toxins 2020, 12, 402. [Google Scholar] [CrossRef]
- Baumann, K.; Casewell, N.R.; Ali, S.A.; Jackson, T.N.W.; Vetter, I.; Dobson, J.S.; Cutmore, S.C.; Nouwens, A.; Lavergne, V.; Fry, B.G. A Ray of Venom: Combined Proteomic and Transcriptomic Investigation of Fish Venom Composition Using Barb Tissue from the Blue-Spotted Stingray (Neotrygon kuhlii). J. Proteom. 2014, 109, 188–198. [Google Scholar] [CrossRef]
- Flaherty, M.M.; Palmer, O.R.; Diaz, J.A. Galectin-3 in Venous Thrombosis: A Possible New Target for Improved Patient Care. Res. Pract. Thromb. Haemost. 2018, 2, 399–400. [Google Scholar] [CrossRef]
- Kirchhoff, K.N.; Billion, A.; Voolstra, C.R.; Kremb, S.; Wilke, T.; Vilcinskas, A. Stingray Venom Proteins: Mechanisms of Action Revealed Using a Novel Network Pharmacology Approach. Mar. Drugs 2022, 20, 27. [Google Scholar] [CrossRef]
- Criscitiello, M.F. What the Shark Immune System Can and Cannot Provide for the Expanding Design Landscape of Immunotherapy. Expert Opin. Drug Discov. 2014, 9, 725–739. [Google Scholar] [CrossRef] [PubMed]
- Gopalan, T.K.; Gururaj, P.; Gupta, R.; Gopal, D.R.; Rajesh, P.; Chidambaram, B.; Kalyanasundaram, A.; Angamuthu, R. Transcriptome Profiling Reveals Higher Vertebrate Orthologous of Intra-Cytoplasmic Pattern Recognition Receptors in Grey Bamboo Shark. PLoS ONE 2014, 9, e100018. [Google Scholar] [CrossRef]
- Xu, X.H.; Lv, P.F.; Wang, T.X.; Wang, B.X.; Shi, Y.; Wang, B.X.; Meng, Z.R.; Chen, Q.X.; Zhuang, J.X.; Wang, Y.Y. Bone-Strengthening Effects and Safety of Compound Peptides from Skin of Chiloscyllium plagiosum and Mustelus griseus. Food Sci. Nutr. 2020, 8, 1522–1533. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.R.; Sampaio, E.; Santos, C.; Couto, A.; Pegado, M.R.; Diniz, M.; Munday, P.L.; Rummer, J.L.; Rosa, R. Absence of Cellular Damage in Tropical Newly Hatched Sharks (Chiloscyllium plagiosum) under Ocean Acidification Conditions. Cell Stress Chaperones 2018, 23, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chen, M.; Wang, X.; Kang, S.; Xue, W.; Li, Z. Identification of Anti-TNFα VNAR Single Domain Antibodies from Whitespotted Bambooshark (Chiloscyllium plagiosum). Mar. Drugs 2022, 20, 307. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wang, M.; Xiang, H.; Jiang, Y.; Gong, J.; Su, D.; Al Azad, M.A.R.; Dong, H.; Feng, L.; Wu, J.; et al. Bamboo Shark as a Small Animal Model for Single Domain Antibody Production. Front. Bioeng. Biotechnol. 2021, 9, 1177. [Google Scholar] [CrossRef] [PubMed]
- Bakke, F.K.; Gundappa, M.K.; Matz, H.; Stead, D.A.; Macqueen, D.J.; Dooley, H. Exploration of the Nurse Shark (Ginglymostoma cirratum) Plasma Immunoproteome Using High-Resolution LC-MS/MS. Front. Immunol. 2022, 13, 873390. [Google Scholar] [CrossRef] [PubMed]
- Raithaus, L.R. Shark Liver Extract for Stimulating the Immune System. U.S. Patent 5840342A, 24 November 1998. [Google Scholar]
- Huang, F.J.; Wu, W.T. Purification And Characterization Of A New Peptide (S-8300) From Shark Liver. J. Food Biochem. 2010, 34, 962–970. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Chen, Y.; Lu, C.; Qian, Y.; Lv, Z. Preliminary Profiling of MicroRNA in the Normal and Regenerating Liver of Chiloscyllium plagiosum. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 24, 60–67. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Chen, J.; Zhang, W.; Sheng, Q.; Chen, J.; Yu, W.; Nie, Z.; Zhang, Y.; Wu, W.; et al. A Shark Liver Gene-Derived Active Peptide Expressed in the Silkworm, Bombyx Mori: Preliminary Studies for Oral Administration of the Recombinant Protein. Mar. Drugs 2013, 11, 1492–1505. [Google Scholar] [CrossRef]
- Oliver, S.P.; Turner, J.R.; Gann, K.; Silvosa, M.; D’Urban Jackson, T. Thresher Sharks Use Tail-Slaps as a Hunting Strategy. PLoS ONE 2013, 8, 67380. [Google Scholar] [CrossRef] [PubMed]
- Viducic, K.; Natanson, L.J.; Winton, M.V.; Humphries, A. Reproductive Characteristics for the Blue Shark (Prionace glauca) in the North Atlantic Ocean. Fish. Bull. 2022, 120, 26–38. [Google Scholar] [CrossRef]
- Moreira, I.; Figueiredo, I.; Farias, I.; Lagarto, N.; Maia, C.; Robalo, J.; Moura, T. Growth and Maturity of the Lesser-Spotted Dogfish Scyliorhinus canicula (Linnaeus, 1758) in the Southern Portuguese Continental Coast. J. Fish Biol. 2022, 100, 315–319. [Google Scholar] [CrossRef] [PubMed]
- McComb, D.M.; Tricas, T.C.; Kajiura, S.M. Enhanced Visual Fields in Hammerhead Sharks. J. Exp. Biol. 2009, 212, 4010–4018. [Google Scholar] [CrossRef]
- Vehniäinen, E.R.; Ruusunen, M.; Vuorinen, P.J.; Keinänen, M.; Oikari, A.O.J.; Kukkonen, J.V.K. How to Preserve and Handle Fish Liver Samples to Conserve RNA Integrity. Environ. Sci. Pollut. Res. 2019, 26, 17204–17213. [Google Scholar] [CrossRef]
- Cavanagh, R.D.; Camhi, M.; Burgess, G.H.; Cailliet, G.M.; Fordham, S.V.; Simpfendorfer, C.A.; Musick, J.A. Sharks, Rays and Chimaeras: The Status of the Chondrichthyan Fishes; Folwer, S.L., Ed.; IUCN: Gland, Switzerland, 2005; ISBN 2831707005. [Google Scholar]
- Smart, J.J.; Harry, A.V.; Tobin, A.J.; Simpfendorfer, C.A. Overcoming the Constraints of Low Sample Sizes to Produce Age and Growth Data for Rare or Threatened Sharks. Aquat. Conserv. 2013, 23, 124–134. [Google Scholar] [CrossRef]
- Skomal, G.B. Shark Nursery Areas in the Coastal Waters of Massachusetts. Am. Fish. Soc. Symp. 2007, 50, 17–33. [Google Scholar]
- Feldheim, K.A.; Gruber, S.H.; Ashley, M.V. The Breeding Biology of Lemon Sharks at a Tropical Nursery Lagoon. Proc. R. Soc. B Biol. Sci. 2002, 269, 1655–1661. [Google Scholar] [CrossRef] [PubMed]
- Wosnick, N.; Niella, Y.; Hammerschlag, N.; Chaves, A.P.; Hauser-Davis, R.A.; da Rocha, R.C.C.; Jorge, M.B.; de Oliveira, R.W.S.; Nunes, J.L.S. Negative Metal Bioaccumulation Impacts on Systemic Shark Health and Homeostatic Balance. Mar. Pollut. Bull. 2021, 168, 112398. [Google Scholar] [CrossRef] [PubMed]
- Smale, M.J.; Jones, R.T.; Correia, J.P.; Henningsen, A.D.; Crow, G.L.; Garner, R. Chapter 39 Research on Elasmobranchs in Public Aquariums. In The Elasmobranch Husbandry Manual; Ohio Biological Survey Columbus: Columbus, OH, USA, 2004; pp. 533–541. ISBN 0867271523. [Google Scholar]
- Tiktak, G.P.; Butcher, D.; Lawrence, P.J.; Norrey, J.; Bradley, L.; Shaw, K.; Preziosi, R.; Megson, D. Are Concentrations of Pollutants in Sharks, Rays and Skates (Elasmobranchii) a Cause for Concern? A Systematic Review. Mar. Pollut. Bull. 2020, 160, 111701. [Google Scholar] [CrossRef]
- Bezerra, M.F.; Lacerda, L.D.; Lai, C.T. Trace Metals and Persistent Organic Pollutants Contamination in Batoids (Chondrichthyes: Batoidea): A Systematic Review. Environ. Pollut. 2019, 248, 684–695. [Google Scholar] [CrossRef]
- Marques, A.F.S.; Alves, L.M.F.; Moutinho, A.; Lemos, M.F.L.; Novais, S.C. Scyliorhinus Canicula (Linnaeus, 1758) Metal Accumulation: A Public Health Concern for Atlantic Fish Consumers? Mar. Pollut. Bull. 2021, 169, 112477. [Google Scholar] [CrossRef] [PubMed]
- Alves, L.M.F.; Nunes, M.; Marchand, P.; Le Bizec, B.; Mendes, S.; Correia, J.P.S.; Lemos, M.F.L.; Novais, S.C. Blue Sharks (Prionace glauca) as Bioindicators of Pollution and Health in the Atlantic Ocean: Contamination Levels and Biochemical Stress Responses. Sci. Total Environ. 2016, 563–564, 282–292. [Google Scholar] [CrossRef]
- Evans, T.G.; Hofmann, G.E. Defining the Limits of Physiological Plasticity: How Gene Expression Can Assess and Predict the Consequences of Ocean Change. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-Throughput Sequencing Technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef]
- Feder, M.E.; Walser, J.C. The Biological Limitations of Transcriptomics in Elucidating Stress and Stress Responses. J. Evol. Biol. 2005, 18, 901–910. [Google Scholar] [CrossRef]
- Teffer, A.K.; Hinch, S.; Miller, K.; Jeffries, K.; Patterson, D.; Cooke, S.; Farrell, A.; Kaukinen, K.H.; Li, S.; Juanes, F. Cumulative Effects of Thermal and Fisheries Stressors Reveal Sex-Specific Effects on Infection Development and Early Mortality of Adult Coho Salmon (Oncorhynchus kisutch). Physiol. Biochem. Zool. 2019, 92, 505–529. [Google Scholar] [CrossRef]
- Henderson, C.J.; Stevens, T.F.; Lee, S.Y. Assessing the Suitability of a Non-Lethal Biopsy Punch for Sampling Fish Muscle Tissue. Fish Physiol. Biochem. 2016, 42, 1521–1526. [Google Scholar] [CrossRef] [PubMed]
- Gleason, L.U. Applications and Future Directions for Population Transcriptomics in Marine Invertebrates. Curr. Mol. Biol. Rep. 2019, 5, 116–127. [Google Scholar] [CrossRef]
- Nachtigall, P.G.; Grazziotin, F.G.; Junqueira-De-Azevedo, I.L.M. MITGARD: An Automated Pipeline for Mitochondrial Genome Assembly in Eukaryotic Species Using RNA-Seq Data. Brief. Bioinform. 2021, 22, bbaa429. [Google Scholar] [CrossRef] [PubMed]
- Feutry, P.; Kyne, P.; Pillans, R.; Chen, X.; Marthick, J.; Morgan, D.; Grewe, P. Whole Mitogenome Sequencing Refines Population Structure of the Critically Endangered Sawfish Pristis pristis. Mar. Ecol. Prog. Ser. 2015, 533, 237–244. [Google Scholar] [CrossRef]
- Fang, J.; Xu, C.; Li, Q. Transcriptome Analysis of Inbreeding Depression in the Pacific Oyster Crassostrea Gigas. Aquaculture 2022, 557, 738314. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).