Abstract
In this study, the complete mitochondrial genomes of the Mexican golden trout, Oncorhynchus chrysogaster, and Nelson’s trout, O. mykiss nelsoni, were assembled and characterized. The mitogenomes were 16,655 bp and 16,661 bp long, respectively, and were composed of 13 protein-coding genes (PCGs), two ribosomal RNA genes, and 22 transfer RNA genes (all with typical ‘cloverleaf’ secondary structures). The length of the D-loop regions was among the longest found in Salmonids, and mitochondrial synteny in both species was identical to that reported in other Salmonids. Selective pressure analysis in the PCGs indicated that purifying selection, mainly among cox and nd genes families, likely generated the main differences between the two studied species. Nine tRNA genes showed slight differences relative to other O. mykiss subspecies, which were identical between the two study taxa. The origin of the light-strand replication has a loop that was especially large in O. mykiss nelsoni. Phylogenetic analysis indicated that O. chrysogaster and O. mykiss nelsoni are sister species, contrary to the expectation that O. chrysogaster would cluster with O. gilae. As previous studies have suggested, O. chrysogaster and O. mykiss nelsoni share common ancestry with North American trout species.
1. Introduction
In past decades, it was common to use short mitochondrial gene fragments to resolve phylogenetic relationships at different taxonomic levels. Recently, whole mitochondrial genome studies have been proposed as a valuable tool for phylogenetic inferences and the modeling of genome evolution [1]. These phylomitogenomic analyses have also been used to resolve a persistent debate about higher-level relationships among teleost fishes or lower-level relationships within species complexes [2,3,4]. For the family Salmonidae, phylogenetic relationships have been reconstructed to understand the evolutionary history of behavioral, life history, and ecological traits [3,5]. Phylogenies using mitogenomes from each of the genera of the family Salmonidae have recently been published [6,7,8,9,10,11,12,13,14,15,16,17,18,19]. This family includes three subfamilies: Salmoninae (salmons, trouts, and charrs), Coregoninae (ciscoes and whitefishes), and Thymallinae (graylings) [20]. Salmonid fish are commercially important, widely used in aquaculture, and have been introduced in 83 countries [21,22]. Consequently, there is great interest in the genetic features of mitochondrial DNA, which can be informative for stock identification, management, conservation, and population studies [3,5,23,24]. The genus Oncorhynchus and five other genera (Brachymystax, Parahucho, Hucho, Salmo, and Salvelinus) comprise the subfamily Salmoninae [20].
In Mexico, only the genus Oncorhynchus Suckley [25] (Actinopterygii: Salmoniformes: Salmonidae) has native representatives with a wide distribution [26,27,28]. However, only two species were formally recognized: the Mexican golden trout, O. chrysogaster, from the Sierra Madre Occidental (SMO) [26,29]; and Nelson’s trout, O. mykiss nelsoni. Nelson’s trout is the southernmost rainbow trout subspecies, and its distribution is restricted to the Baja California Peninsula. This subspecies is slow-growing, short-lived, rapidly maturing, and reaches a smaller length compared to the subspecies O. m. mykiss (used typically in hatcheries). Traditional and genetic data have established Nelson’s trout as a subspecies of O. mykiss corresponding to a distinct and native genetic group [30,31,32,33,34]. However, comparatively little is known about Mexican golden trout (although see [26,27,29]). The rest of Mexico’s native trout biodiversity represents a complex of undescribed species, which contains an undetermined number of new species that inhabit the SMO to the north and south of where the Mexican golden trout is distributed; these taxa are currently referred to by the name of the watershed where they live [26,28,35,36,37,38,39,40,41,42,43,44,45,46]. Despite the presence of these native species, the rainbow trout O. mykiss has been introduced for farming in natural aquatic habitats of the SMO [26,27,28,39] and most other Mexican mountain ranges [26,28]. In the case of the SMO, such introductions have led to hybridization between Mexican native trout and rainbow trout [40,42,43].
The species radiation of the genus Oncorhynchus was promoted by geological activity, climatic changes, and geographic isolation, which led them to evolve in highly dynamic environments [44]. The diversification of this genus has been widely discussed and molecular dating varies depending on the molecular marker employed (SNPs [47], mitochondrial genes [48], nuclear genes [49] and mitonuclear genes [50]). However, there is general agreement that the origin of Pacific salmons and Pacific trouts occurred during the Miocene. The Pacific trouts include O. mykiss, O. clarkii, O. gilae, O. apache (now O. g. apache) and the SMO’s undescribed trout complex [33,50]. Subsequently, radiation into the O. mykiss and O. clarkii complexes occurred during the Pleistocene [51,52] or possibly earlier during the Late Pliocene [50]. Protein polymorphism analysis established that O. gilae and O. gilae apache are sister taxa that, together with the Mayo trout (an undescribed northern species from the SMO), are more closely related to the O. mykiss complex than to the O. clarkii complex, which supports the divergence hypothesis proposed by Loudenslager et al. [53]. In addition, the Yaqui trout from SMO (another undescribed northern species) showed a closer phylogenetic relationship to the O. mykiss complex, which included O. mykiss nelsoni [54,55]. Lately, Mayden et al. [28] explored these phylogenetic relationships with an increased sampling effort that included the undescribed trout complex of the SMO and the O. gilae lineage. They found that only the Southern Conchos trout (on the Atlantic slope) clustered into the O. gilae lineage, while the other trout from a different watershed of the SMO (on the Pacific slope) showed multiple divergent lineages that were more closely related to the O. mykiss lineage than to the O. gilae lineage. For this reason, exploring phylogenetic relationships with a powerful tool like mitogenomes could help to resolve the numerous taxonomic uncertainties in these species complexes.
In this work, we characterized for the first time the complete mitochondrial genome of the two recognized Mexican trout taxa and used only the mitochondrial protein-coding genes (PCGs) to clarify their genetic relationships within the genus Oncorhynchus. We have assembled and characterized the whole mitogenome and annotated its genes for O. chrysogaster and O. mykiss nelsoni. Additionally, we assessed the phylogenetic position of the two species of interest using the mitochondrial PCGs of the Oncorhynchus genus available from NCBI GenBank. The information in this work will form the basis of future genetic studies of Mexican trouts and could provide critical information for managing these native species.
2. Methods
Adult Oncorhynchus chrysogaster (Needham & Gard [29]) were collected from the Fuerte River, Chihuahua, Mexico using a Smith Root 15-B POW (110V AC) electrofishing rig and kept in captivity at the Aquaculture Center of INAPESCA, Guachochi, Chihuahua, Mexico. An individual of Oncorhynchus mykiss nelsoni (Evermann [30]) was collected from a stream segment located between Rancho Mike’s Sky and Rancho Garet in San Pedro Mártir, Baja California, Mexico, with a Smith-Root model LR24 backpack electrofisher and transported to Centro de Investigación Científica y de Educación Superior de Ensenada (CICESE, Ensenada, BC, MEX) and kept in recirculating systems until sampling. The fishes analyzed were rapidly and humanely killed. The complete specimen was fixed in ethanol. About 15 mg of fin tissue was used for total DNA extraction with the DNeasy Blood & Tissue Kit (QIAGEN®, Hilden, NRW, DEU) and quantified using a spectrophotometer (Nanodrop 2000®; Thermo Scientific, Wilmington, DE, USA). Total DNA (1000 ng/sample) was sent to the Genomics Center of Georgia (University of Georgia, Athens, GA, USA) for genomic sequencing. The DNA was sheared with a Bioruptor® device by sonication using two rounds of five cycles alternating 30 s of sonication with 30 s without sonication on the high setting. The protocol for library preparation was followed using the Kapa Biosystems® Hyper Prep Kit (KR0961–v4.15), ligating custom adapter and amplifying with 12 cycles of PCR with custom nucleotide indexed primers [56]. Magnetic beads (Speed Beads) were used for Dual-size selection, performed to recover a fragment of ~250–450 bp in size [57]. Libraries were sequenced to produce paired-end 150-nucleotide reads in an Illumina HiSeq 4000 at the Oklahoma Medical Research Foundation Clinical Genomics Center (Oklahoma City, OK, USA).
Fastq files were cleaned by trimming low-quality regions of the reads with CLC Genomics Workbench 10.1.1 software using the default parameters (Trim using quality scores, limit = 0.05, Phred value equivalent to 13.01, and Trim ambiguous nucleotides, number of ambiguities max = 2), and followed by de novo assembly with the following settings: automatic bubble size (50) and word size (20), minimum contig length (200 bp), perform scaffolding (yes), and auto-detect paired distances. The longest contig identified was used as a query to search the nucleotide NCBI GenBank [58] database using the Basic Local Alignment Search Tool (Blast) [59].
To annotate and characterize the complete mitochondrial genome, we followed the recommendations of Baeza [60]. The assembled mitochondrial genomes of O. chrysogaster and O. mykiss nelsoni were annotated using the web servers MITOS and MITOS2 [61,62] using the mitochondrial vertebrate genetic code. The in silico annotated mitochondrial genomes were curated manually, considering start and stop codons corrections in the web server ExPASy [63], and MEGA X [64]. The manually curated annotations were compared with other salmonid mitochondrial genomes in the NCBI nucleotide database (O. mykiss-NC_001717, O. clarkii virginalis-MW300344, O. clarkii henshawi-AY886762, O. keta-AP010773, and O. gilae-MW300342). The visualizations of the mitogenomes were performed with the web server GenomeVx [65].
Nucleotide composition and codon usage profiles were analyzed for the PCGs in the two mitochondrial genomes. The nucleotide composition was estimated in the software MEGA X for each mitochondrial sequence [64] and compared with the composition of all mitogenomes of the family Salmonidae available in the NCBI nucleotide database [58] (Table S1). Codon usage was estimated in the Sequence Manipulation Suite (SMS) web server using the vertebrate mitochondrial code [66]. Relative synonymous codon usage (RSCU) of concatenated PCGs for each species was visualized using the EZcodon tool in the web server EZmito [67].
The substitution rates of the different PCGs were estimated between closely related species. The values of KA = dN = SA/LA (the number of non-synonymous substitutions per non-synonymous site), KS = dS = SS/LS (number of synonymous substitutions per synonymous site) and ω (the KA/KS ratio) were evaluated with the software KAKS_Calculator2.0 [68]. The ω is a measure of the selective pressures acting on each gene. Values of ω = 1 indicates neutrality, while ω < 1 indicates negative or purifying selection, and ω > 1 indicates positive or diversifying selection [68]. The ω values were calculated by pairwise comparison between O. chrysogaster and O. masou biwa (GenBank: EF105342) and between O. mykiss nelsoni and O. masou biwa (GenBank: EF105342). Oncorhynchus masou biwa was selected because it is the congener species that allowed estimation of the substitution rates for almost all the analyzed genes. Other candidate congeners were also tested (O. clarkii, O. gilae and O. mykiss), but due to their phylogenetic closeness, it was not possible to estimate non-synonymous substitution rates for any of the genes (results not shown). The γ-MYN model [69] accounted for variable mutation rates at sequence sites. Finally, significant differences in ω between species (p < 0.05) were calculated using the KAKS_Calculator2.0 software to indicate either negative (purifying) selection or positive (diversifying) selection.
Transfer RNAs genes (tRNA) were identified using the software MiTFi [70] implemented in the web server MITOS. The tRNA’s secondary structure was visualized for each gene with the tool Forna in the ViennaRNA web server [71].
The control region (CR) was also further examined for the two studied mitochondrial genomes. The putative CR’s nucleotide composition was analyze using MEGA X software [64].
PCG-based phylogenetic analysis can be helpful for resolving the phylogenetic relationships among salmonids [13,16]. In addition, an artifact of node overestimation is known to occur when the entire mitogenome is used [13,16]. For this reason, we explored the phylogenetic position of O. chrysogaster and O. mykiss nelsoni relative to other representative species of the genus Oncorhynchus based on their PCGs. The concatenated PCG sequences were aligned using CLUSTAL OMEGA [72], then the best-fitting nucleotide substitution model was selected with JMODELTEST2 [73,74]. Akaike (AIC) and Bayesian (BIC) information criteria were used with default settings. The phylogenetic analysis was conducted using MRBAYES 3.2 [75] with the general time-reversible model (GTR) using a gamma-distributed rate variation among sites (+G) and proportion of invariable sites (+I). Markov chain Monte Carlo simulations were run with 10,000 generations, 100 sample frequencies, and default settings. The phylogenetic tree was visualized using FIGTREE V1.4.4 [76]. This analysis was performed using the two newly sequenced and annotated mitogenomes of O. chrysogaster and O. mykiss nelsoni, together with those of other species within the genus and five species as outgroups (Parahucho perryi, Salmo salar, Salvelinus fontinalis, S. leucomaenis, S. levanidovi), which were downloaded from the NCBI web server (Table S2).
3. Results and Discussion
After trimming, 5,578,930 reads were obtained for O. chrysogaster with 140 bp of average length, which produced 24,353 contigs with 340 bp of average length (N50 = 447). In comparison, 6,614,455 read were obtained for O. mykiss nelsoni with 138 bp of average length, which produced 22,913 contigs with 357 bp of average length (N50 = 472). The mitochondrial genomes of the Mexican golden trout O. chrysogaster (OP902890) and the Nelson trout O. mykiss nelsoni (OP902891) were assembled and circularized with a coverage of 21.69 X and 43.97 X, respectively.
The Mexican trouts, O. chrysogaster and O. mykiss nelsoni, had similar total mitochondrial genome lengths—16,655 bp and 16,661 bp, respectively. In both taxa, the mitochondrial genomes were compact, and few intergenic spaces and overlaps among gene junctions were found (Figure 1, Table 1). In both species, the mitochondrial genome comprised 13 PCGs, two ribosomal RNA genes (rrnS [12S] and rrnL [16S]), and 22 transfer RNA genes (tRNA) (Table 1). A single long intergenic space was assumed to be the CR, with a length of 1002 bp in O. chrysogaster and 1004 bp in O. mykiss nelsoni. Three additional short intergenic spaces were observed in the mitochondrial genome, ranging from 14 to 36 bp in each species (21 bp between tRNA-Val and 16S, ol-31 bp, 14 bp between tRNA-Asp and cox1 [O. chrysogaster]; 21 bp between tRNA-Val and 16S, ol-36 bp, 14 bp between tRNA-Asp and cox1 [O. mykiss nelsoni]) (Table 1).
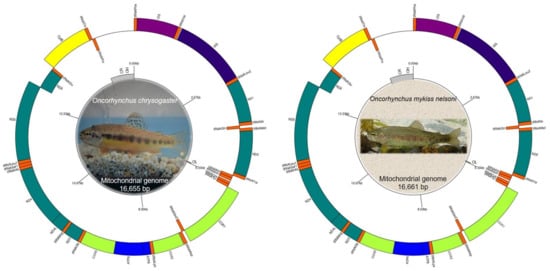
Figure 1.
Circular mitochondrial genome maps of the Mexican golden trout, Oncorhynchus chrysogaster, and Nelson’s trout, O. mykiss nelsoni. The annotated maps depict 13 protein-coding genes (PCGs), two ribosomal RNA genes (rrnS-12S and rrnL-16S) and 22 transfer RNA (tRNA) genes in the external circle. The external or inner positioning in the circle indicates whether the gene was encoded on the heavy strand (+) or the light strand (−), respectively. The putative control region and the origins of replication were indicated in gray in the innermost circle of the map. Photo credit: O. chrysogaster photo by FJ García-De León; O. mykiss nelsoni photo by Ivan Alejandro Meza Matty (Doi: 10.13140/RG.2.2.36230.86080).

Table 1.
Gene arrangement of the mitochondrial genome of the Mexican golden trout, Oncorhynchus chrysogaster, and Nelson’s trout, O. mykiss nelsoni.
In both species, most (n = 12) of the PCGs, 14 tRNA, and the two rRNA genes were encoded on the heavy strand; only the nd6 gene and the remaining eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser2, tRNA-Glu, tRNA-Pro) were encoded on the light strand (Figure 1, Table 1). The same pattern occurs in Salmo ischchan [15], hybrids of O. mykiss x Atlantic salmon [77], and in eight other families of bony fishes (Acipenseridae, Catostomidae, Percidae, Clupeidae, Centrarchidae, Polyodontidae, Cyprinidae, Ictaluridae) [78].
The mitochondrial gene arrangement observed was identical among the 14 species and subspecies of trouts of the genus Oncorhynchus, including the study taxa O. chrysogaster and O. mykiss nelsoni. Furthermore, mitochondrial gene arrangement in this genus was identical to that reported in other Salmonids (e.g., Brachymystax lenok-JQ686730, Coregonus lavaretus-MK913369, Hucho bleekeri-HM804473, Prosopium cylindraceum-JQ390062, Salmo trutta-MF621763, Salvelinus albus-KT266871, Salvethymus svetovidovi-MK695627, Stenodus leucichthys-JQ390059, Thymallus thymallus-MT410870). The only difference in the gene arrangement was in oh and ol regions, which are frequently not reported. This is similar to findings by Si et al. [79] that the gene content, genes arrangement, and base composition of two Lenoks (Brachymystax spp.) were nearly identical to most other teleosts. Similarly, Levin et al. [15] considered the gene arrangement of Salmo spp. to be similar to other vertebrates. The mitochondrial gene arrangement in the genus Oncorhynchus is therefore not a distinguishing characteristic for differentiation among species, as has been reported in some other fishes, such as Chirostoma humboldtianum [80] and Anoplopoma fimbria [81], as well as octopuses [82] and some gastropods of the Strombidae family [83]. This contrasts with other taxa, such as gastropods of the Vermetidae family [83] or anomuran crustaceans [84], in which gene arrangement is relevant for the differentiating species or genera.
The total nucleotide composition of the entire mitochondrial genome in O. chrysogaster was as follows: 27.9% A, 26.2% T, 29.0% C, and 17.0% G, resulting in a 54.1% AT-content and a 45.9% GC-content. In O. mykiss nelsoni, the composition was: 27.9% A, 26.1% T, 29.0% C, and 17.0% G, for a 54.1% AT-content and a 45.9% GC-content. Overall, the AT-content detected in the two mitochondrial genomes is within the range described for other salmonid fishes. Thymallus arcticus exhibits the highest AT content reported, with 55.9%, while Coregonus autumnalis, C. clupeaformis, C. lavaretus, C. ussuriensis, and C. peled, have the lowest reported AT-content (52.4%) (Table S1).
The PCGs in the mitochondrial genome of O. chrysogaster and O. mykiss nelsoni comprise 11,430 nucleotides corresponding to 3810 total codons in both cases. For all but one of the PCGs, the start codon was ATG (12 PCGs: nd1, nd2, cox2, atp8, atp6, cox3, nd3, nd4l, nd4, nd5, nd6, cytb). The exception was cox1, whose start codon was GTG. The most frequent stop codon was TAA (six PCGs: cox1, atp8, atp6, cox3, nd4l, nd5); three PCGs used TAG (nd1, nd3, nd6); and four PCG presented a truncated stop codon T (nd2, cox2, nd4, cytb) (Table 1). The same pattern has been reported for O. mykiss [85], O. tschawytscha [24], Hucho taimen [86], Brachymystax lenok tsinlingensis [87], hybrids of O. mykiss × Atlantic salmon [77], Thymallus arcticus grubei [88], and Salmo ischchan [15]. The incomplete stop codon, common in the mitogenomes of teleost fishes, is completed later on via posttranscriptional polyadenylation [24,77,85,86,87].
For the PCGs, the relative synonymous codon usage (RSCU) and amino acid composition are summarized in Figure 2 for O. chrysogaster and O. mykiss nelsoni. In O. chrysogaster, the most frequently used codons (amino acids) were CTA (Leu), which was used 184 times (28%); CTT (Leu), used 152 times (23%); GCC (Ala) used 151 times (44%); and CTC (Leu) used 146 times (22%). The least commonly used codons (amino acids), excluding stop codons, included TGT (Cys) used eight times (30%), followed by CGG and CGT (Arg) used eight times each (11%), and TCG (Ser) used nine times (4%). Meanwhile, in O. mykiss nelsoni, the most frequently used codons were CTA (Leu) used 183 times (28%); GCC (Ala) used 151 times (44%); CTT (Leu) used 151 times (23%); and CTC (Leu) used 150 times (23%). The least common were TGT (Cys) used eight times (3%); and CGG and CGT (Arg) used eight times each (11%). The RSCU and amino acid composition have been reported for PCGs in the salmonids Brachymystax lenok tsinlingensis and Salvelinus malma, in which the most frequent amino acid was Leu and the least common was Cys [87,89]. CTT was one of the most used codons, similar to other vertebrates; however, unlike other vertebrates, AAG is not the least-used codon [89,90].
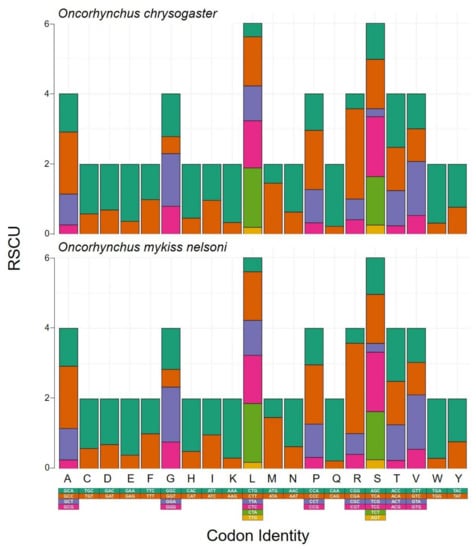
Figure 2.
Relative synonymous codon usage (RSCU) in the Mexican golden trout, Oncorhynchus chrysogaster, and Nelson’s trout, O. mykiss nelsoni. Letters represent the single letter abbreviation name of each amino acid.
The mitochondrial PCGs exhibited KA/KS ratio values <0.057 (p-value < 0.05 in all cases) for O. chrysogaster and O. mykiss nelsoni. In O. chrysogaster, the nd6 gene had the highest KA/KS value (0.057), followed by the nd5 and nd2 genes (0.047 and 0.045, respectively). In contrast, the atp8, cox family genes, nd4 and cytb showed zero or near zero values (Figure 3). In O. mykiss nelsoni, the nd5 gene had the highest value (0.054), followed by nd6 –0.048, atp8–0.046 and nd2–0.044. In contrast, again, the values for cox1, cox3 and cytb were closest to zero (Figure 3). The KA/KS values in these two species were more similar to those of the freshwater salmonids than to anadromous salmonids [19]. In general, the observed KA/KS values were lower than other fishes, such as cichlids (<0.143) [91] or Cobitinae (<0.120) [92]. These results indicate that all mitochondrial PCGs from both trouts analyzed are under purifying selection, which is higher in some cases, such as the cox and cytb genes. On the other hand, it is common for the atp gene to show more relaxed selection than cox family genes in vertebrates [93]; for this reason, the cox genes are potential barcoding markers [92].
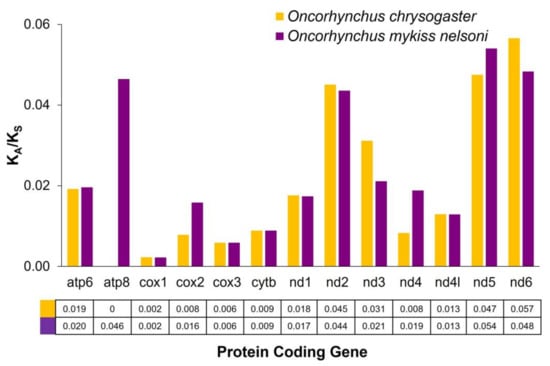
Figure 3.
Selective pressure analysis of the protein-coding genes of the Mexican golden trout, Oncorhynchus chrysogaster, and Nelson’s trout, O. mykiss nelsoni.
In the mitochondrial genome of O. chrysogaster and O. mykiss nelsoni, the length range of 22 tRNA genes was between from 67 bp to 75 bp. The 22 tRNA genes exhibited a typical ‘cloverleaf’ secondary structure (Figure 4), except the tRNA-Ser1, which has a truncated dihydroxyuridine (DHU) loop. This tRNA-Ser lost the DHU arm, and the nucleotides formed a simple loop. This loss is a common trait among metazoan mitogenomes, although it is not clear if it is reflected in a modification in their functions; mismatched base pairs are considered as irreversible, evolutionary-derived states, which might be caused by tRNA editing, as reported in Cobitinae and hybridized Salmonids [77,92]. However, O. mykiss and O. tschawytscha are an exception to this pattern because they have a complete DHU arm [24,85]. A similar pattern was reported in Gadus morhua and Brachymystax lenok tsinlingensis, except they also present a large anticodon stem [87,94]. Additionally, both trout species studied here showed slight differences from the previously described mitochondrial genome of O. mykiss in nine tRNA genes (tRNA-Arg, tRNA-Asp, tRNA-Cys, tRNA-Glu, tRNA-Leu1, tRNA-Leu2, tRNA-Ser1, tRNA-Trp, tRNA-Val). Two tRNA genes had one substitution (a transition and a transversion), four had an insertion of 1–2 nucleotides, and three had deletions of 1–3 nucleotides.
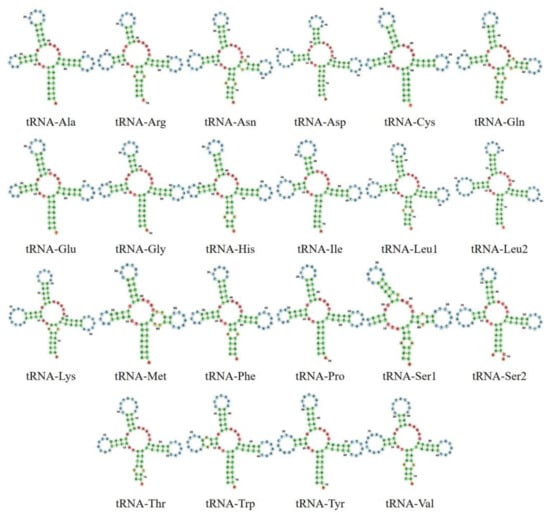
Figure 4.
Secondary structures of 22 transfer RNA genes in the Mexican golden trout, Oncorhynchus chrysogaster, and Nelson’s trout, O. mykiss nelsoni. All the secondary structures of the transfer RNA were identical between the two species, so are only shown once.
The 12S rRNA gene length in O. chrysogaster and O. mykiss nelsoni was 947 pb with an AT-content of 49.5%. The length of the 16S rRNA gene was 1659 and 1658 pb, with an AT-content of 51.7 and 51.6% in each species, respectively, due to one G deletion in O. mykiss nelsoni. Furthermore, as in other Salmonids, they were in the H-strand between tRNA-Phe and tRNA-Leu2 and separated by tRNA-Val. For the rRNAs, the nucleotide usage has rarely been reported in Salmonids (AT-content 50.97%) [87], but the AT-content in these two species was within the range found in other Salmonids (calculated from sequences published in GenBank; see Table S3). The Salmonids with the lowest AT content were Prosopium cylindraceum (12S rRNA = 48.8%), Coregonus clupeaformis, and Stenodus leucichthys (16S rRNA = 51.0%), while the highest values were found for O. masou masou (12S rRNA = 50.5%, 16S rRNA = 52.9%) and Salmo salar (12S rRNA = 50.5%).
In O. chrysogaster and O. mykiss nelsoni, the putative control regions (CR) were 1002 bp and 1004 bp long, respectively, with a high AT content (61.5%). The putative CR was located between the tRNA-Pro and tRNA-Phe genes (Figure 1); this includes the origin of H-strand replication within it. For O. chrysogaster, the CR begins at position 15,654 and ends at position 16,655, while for O. mykiss nelsoni, it starts at position 15,658 and ends at position 16,661. The AT content in the CR estimated of other Salmonids, such as O. masou formosanus is 60.0% [13], and for H. taimen is 61.9% [86]. Our results indicate that the two trouts studied here exhibit an AT content within the range for other species of the family Salmonidae (range: 58.8–66.1% AT, 505–1339 bp long) (Table S4).
Another non-coding region, the origin of light-strand replication (ol) was in a cluster of five tRNA genes (the WANCY region) between tRNA-Asn and tRNA-Cys (Figure 1). The ol comprises 51 nucleotides in O. chrysogaster and 56 in O. mykiss nelsoni; this region can form a secondary structure with a loop of 13 nucleotides in O. chrysogaster and 18 nucleotides in O. mykiss nelsoni. The stem sequence is conserved among vertebrates, whereas the loop sequence is more variable [24,85,87]. Furthermore, there is a highly conserved motif (5′–GCCGG–3′) which is considered important in transcription and regulation in most vertebrates [6,7,13,24,85,86,87]. The rainbow trout ol loop is noted for its large size and the presence of a stretch of cytosine residues, which differs from mammalian mtDNA sequences, in which this region presents a T-rich residues. Loss of this sequence during the evolution of vertebrates emphasizes the tendency of mtDNA towards a compact genome size [85].
In the BI tree, O. mykiss and O. mykiss nelsoni clustered into a single strongly supported clade (100%) that includes hybrids of O. mykiss x Salmo salar (Figure 5). Oncorhynchus chrysogaster clustered into this clade with 100% support. Meanwhile, O. gilae and O. g. apache grouped together (100%) into a sister clade of O. mykiss and O. chrysogaster. Interestingly, this is the first time that mitochondrial genomes of O. gilae have been included in a phylogenetic analysis. The close relationship between the clades O. mykiss and O. gilae with the monophyletic clade of O. clarkii was strongly supported, consistent with some previous studies using mitochondrial DNA [13,19,48] and other molecular markers [50]. The other cluster includes three clades were monophyletic and highly supported (100 %); O. masou; O. kisutch and O. tshawytscha; and O. gorbuscha, O. keta and O. nerka. The position of this last lineage has been less consistent among studies; in some studies, it has been placed as the most basal clade of the Oncorhynchus genus [8], while in others, it has been placed as an internal clade, close to O. kisutch and O. tshawytscha [3,13,14]. These three monophyletic groups, in turn, form a clade with a high support value (99%). Our results are in concordance with some previous studies [19,48,50], but inconsistent with other phylogenies reconstructed with mitochondrial genomes that found these clades to be independent but weakly supported [3,8,13]. Incorporation of the complete mitogenomes of the different species of the Oncorhynchus genus (including the Mexican native and undescribed species) and the correct outgroups will surely improve the understanding of the phylogenetic relationships of American salmonids.
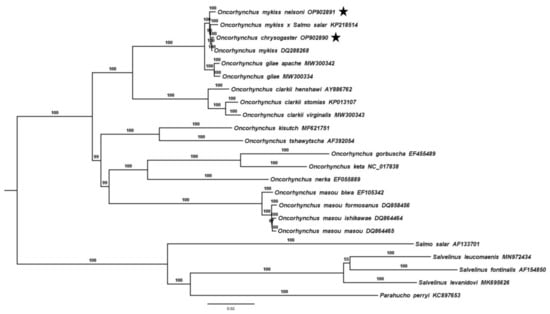
Figure 5.
Phylogenetic tree obtained from the Bayesian inference (BI) analysis based on a concatenated alignment of nucleotides of the 13 protein-coding genes present in the mitochondrial genome of the Mexican golden trout Oncorhynchus chrysogaster, Nelson’s trout O. mykiss nelsoni, and other representatives of the genus Oncorhynchus. The robustness of the BI tree topology was ascertained by 10,000 generations derived from the best-fit model (GTR+G+I) (percent node support is noted above each bar). The number before each species name is the GenBank accession number. Stars mark the mitogenomes obtained in this study.
The origin and diversification of the Mexican trouts have been subjects of controversy and interest for which there are currently three plausible hypotheses that all revolve around their diversification during the glaciations of the Pleistocene [51,53,95,96]. Behnke’s hypothesis [51] sustains that they could be derived from an anadromous trout from the North Pacific that took refuge in the Gulf of California, entering the hydrographic basins that flowed into the Gulf and remaining isolated in the upper areas. This ancestral trout is proposed to have given rise to early radiation, including the taxon that evolved into O. chrysogaster, O. gilae, O. apache (now O. g. apache) and the undescribed trout of the SMO. Therefore, these trout share a common ancestor; this has been supported by the presence of primitive morphological characters shared by Rainbow trout (O. mykiss) and Cutthroat trout (O. clarkii spp.) [51]. A second hypothesis states that an ancestor of the Cutthroat trout moved up the Columbia River basin, then split into two groups. One branch became isolated and evolved into the Yellowstone cutthroat trout, which then moved down the Missouri River, returned, and crossed the Colorado River to the Sierra Mountains. These trout evolved into the California golden trout, which evolved into the Rainbow trout [96]. Finally, the trout forms from Sacramento could then have given rise to the mainland Mexican trout (different from the Rainbow trout group), which shared a common ancestor with the Gila trout inferred by similar characteristics like spotting pattern and coloration [53]. In a third hypothesis, Needham and Gard [29] proposed that Mexican golden, Gila, and Apache trouts could have a hybrid origin from Cutthroat trout and Rainbow trout in the Colorado River basin. However, this hypothesis has been dismissed since it was inconsistent with hybridization experiments [95].
Mayden et al. [28] reported a complete study of Mexican trouts’ relationships to their close relatives using nuclear and mitochondrial genes. They found that O. g. gilae, O. g. apache, and the Mexican trout form a group that is distinct from O. clarkii; similar results were reported in another study where mitochondrial and nuclear genes were also used [50], consistent with the previous hypothesis of a common ancestor between these two groups. Our results with the PCGs also indicate a common ancestor between these groups. Mayden et al. [28] also found a sister relationship between O. g. gilae and O. g. apache and the southern Rio Conchos trout, supporting the close relationship between Gila and the Mexican trout [28]. All these Mexican trouts, which also include the undescribed trouts of the SMO, diverge into multiple independent lineages that are more closely related to O. mykiss from hatcheries than to the Gila lineage [28]. However, the relationship between the Gila lineage and the mainland Mexican trout must be clarified. The mitogenomes assembled de novo in this study support the separation of the Gila trout from the Mexican golden trout. However, the lack of a mitogenome of the Conchos trout makes it impossible to determine whether there is a close relationship between the Gila lineages and other SMO trout. Mayden et al. [28] established that the trout lineage from the SMO is native not a hatchery-reared strain or introduced fish and it constitutes a distinct native lineage from the Mexican golden trout. Therefore, our results support the Behnke hypothesis. However, the adding of mitogenomes from undescribed native Mexican trouts could better clarify the phylogenetic relationships of those SMO trouts.
4. Conclusions
Selective pressure analyses indicated that all mitochondrial PCGs are under purifying selection, which was especially evident for some genes (i.e., cox and cytb). These analyses are rarely conducted on salmonids. A phylogenomic analysis based on a concatenated alignment of nucleotides of PCGs was consistent with O. mykiss nelsoni being a true subspecies of the O. mykiss lineage. In addition, our phylogeny showed a sister relationship between O. chrysogaster and O. mykiss nelsoni, which confirms a common ancestor with North American trouts, as was proposed by Behnke [51]. Furthermore, the analysis found a close relationship between O. mykiss and the Gila trout, O. gilae, although contrary to expectations, O. chrysogaster did not cluster into the same clade as the Gila trout. To fully resolve the relationships and origin of Mexican trouts, mitogenomes of the Conchos trout and others from the SMO will be required.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/fishes8040178/s1, Table S1: Nucleotide composition of the complete mitochondrial genome in Salmonids; Table S2: Species used in the phylogenetic analysis; Table S3: Nucleotide composition of the 12S and 16S rRNA genes in Salmonids; Table S4: Nucleotide composition of the Control Region in Salmonids.
Author Contributions
A.C.: conceptualization, methodology, software, validation, formal analysis, investigation, data curation, writing—original draft preparation, visualization; M.A.D.R.-P.: methodology, resources, data curation, writing—review and editing, supervision, funding acquisition; F.L.-D.l.C.: writing—review and editing, supervision, funding acquisition; G.I.-D.l.M.: writing—review and editing, resources, supervision; F.J.G.-D.L.: conceptualization, resources, writing—review and editing, visualization, supervision, project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by SAGARPA-INAPESCA project number 2505160167, SAGARPA FIRCO-2012 project number GO/04/05/2012-02, and CICESE internal projects: O0F092 and O0F125.
Institutional Review Board Statement
This article does not contain any studies with human participants. All animals used in this study were collected with the permits to fishing (SGPA/DGVS/02485/13, SGPA/DGVS/02968/14 and SGPA/DGVS/05052/15) and issued by the SEMARNAT General Directorate of Wildlife.
Data Availability Statement
Datasets presented in this study can be found in the NCBI online repository (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov (accessed on 1 March 2023)).
Acknowledgments
The first author thanks the Consejo Nacional de Ciencia y Tecnología (CONACyT) for the fellowship (No. 1193058) granted to conduct postdoctoral research. Many thanks Gorgonio Ruiz Campos and Carmen Paniagua Chávez who helped us in the sampling and maintenance of the fishes. Finally, we thank the anonymous reviewers for their comments that improved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hrbek, T.; Farias, I.P. The complete mitochondrial genome of the pirarucu (Arapaima gigas, Arapaimidae, Osteoglossiformes). Genet. Mol. Biol. 2008, 31, 293–302. [Google Scholar] [CrossRef]
- Lavoué, S.; Miya, M.; Inoue, J.; Saitoh, K.; Ishiguro, N.B.; Nishida, M. Molecular systematics of the gonorynchiform fishes (Teleostei) based on whole mitogenome sequences: Implications for higher-level relationships within the Otocephala. Mol. Phylogenet. Evol. 2005, 37, 165–177. [Google Scholar] [CrossRef]
- Horreo, J.L. Revisiting the mitogenomic phylogeny of Salmoninae: New insights thanks to recent sequencing advances. PeerJ 2017, 5, e3828. [Google Scholar] [CrossRef]
- D’Agaro, E.; Gibertoni, P.; Marroni, F.; Messina, M.; Tibaldi, E.; Esposito, S. Genetic and Phenotypic Characteristics of the Salmo trutta Complex in Italy. Appl. Sci. 2022, 12, 3219. [Google Scholar] [CrossRef]
- Yasuike, M.; Jantzen, S.; Cooper, G.A.; Leder, E.; Davidson, W.S.; Koop, B.F. Grayling (Thymallinae) phylogeny within salmonids: Complete mitochondrial DNA sequences of Thymallus arcticus and Thymallus thymallus. J. Fish Biol. 2010, 76, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-Q.; Tong, G.-X.; Bai, Q.-L.; Wang, B.-Q.; Yin, J.-S. The complete mitochondrial genome sequence of white-spotted char (Salvelinus leucomaenis). Mitochondrial DNA 2013, 26, 700–701. [Google Scholar] [CrossRef]
- Zhao, J.-W. The complete mitochondrial genome of the Thymallus grubii (Amur grayling). Mitochondrial DNA 2013, 26, 799–800. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-W.; Chen, J.J.; Lee, T.-H.; Lin, H.-J. Complete mitochondrial genome of Oncorhynchus masou formosanus (Jordan & Oshima, 1919) (Pisces, Salmonidae). Mitochondrial DNA Part B 2016, 1, 295–296. [Google Scholar] [CrossRef]
- Jia, Z.-H.; Xue, S.-Q.; Chen, W.-J.; Zhang, J.-Y. Complete mitochondrial genome of Coregonus cluncaformis. Mitochondrial DNA Part A 2015, 27, 4461–4462. [Google Scholar] [CrossRef]
- Yi-Fan, L.; Huai-Ning, L.; Qi, Z.; Dan, W. Complete mitochondrial genome of Coregonus autumnalis. Mitochondrial DNA Part A 2015, 27, 2498–2499. [Google Scholar] [CrossRef]
- Balakirev, E.S.; Parensky, V.A.; Kovalev, M.Y.; Ayala, F.J. Complete mitochondrial genome of the white char Salvelinus albus (Salmoniformes, Salmonidae). Mitochondrial DNA Part A 2015, 27, 3753–3754. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Jiang, H.; Sun, P.; Chen, J.; Li, L.; Zhang, X.; Yuan, L. Phylogeny and dating of divergences within the genus Thymallus (Salmonidae: Thymallinae) using complete mitochondrial genomes. Mitochondrial DNA Part A 2015, 27, 3602–3611. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Liu, L.-Q.; Guo, B.-Y.; Ye, Y.-Y.; Lü, Z.-M. The complete mitochondrial genome of Oncorhynchus masou formosanus (Salmoniformes: Salmonidae) and phylogenetic studies of Salmoninae. Conserv. Genet. Resour. 2017, 9, 281–284. [Google Scholar] [CrossRef]
- de Flamingh, A.; Mallott, E.K.; Roca, A.L.; Boraas, A.S.; Malhi, R.S. Species identification and mitochondrial genomes of ancient fish bones from the Riverine Kachemak tradition of the Kenai Peninsula, Alaska. Mitochondrial DNA Part B 2018, 3, 409–411. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.; Simonov, E.; Rastorguev, S.; Boulygina, E.; Sharko, F.; Tsygankova, S.; Gabrielyan, B.; Roubenyan, H.; Mayden, R.; Nedoluzhko, A. High-throughput sequencing of the mitochondrial genomes from archived fish scales: An example of the endangered putative species flock of Sevan trout Salmo ischchan. Hydrobiologia 2018, 822, 217–228. [Google Scholar] [CrossRef]
- Arai, Y.; Yokoyama, C.; Nagase, K.; Suwa, M.; Ogawa, Y.; Iuchi, K.; Hisatomi, H. Complete mitochondrial DNA sequences of two endemic subspecies, Salvelinus leucomaenis imbrius and Salvelinus leucomaenis pluvius (Salmonid, White spotted charr) in Japan. Mitochondrial DNA Part B 2019, 4, 1524–1525. [Google Scholar] [CrossRef]
- Oleinik, A.G.; Skurikhina, L.A.; Kukhlevsky, A.D.; Semenchenko, A.A. Complete mitochondrial genome and phylogenetic position of the Taranetz charr Salvelinus taranetzi Kaganovsky, 1955 (Salmoniformes: Salmonidae). Mitochondrial DNA Part B 2019, 4, 2491–2492. [Google Scholar] [CrossRef]
- Oleinik, A.G.; Skurikhina, L.A.; Kukhlevsky, A.D.; Semenchenko, A.A. Complete mitochondrial genome and phylogenetic position of the Levanidov’s charr Salvelinus levanidovi Chereshnev, Skopetz et Gudkov, 1989 (Salmoniformes, Salmonidae). Mitochondrial DNA Part B 2020, 5, 2514–2515. [Google Scholar] [CrossRef]
- Wang, Y.; Xiong, F.; Song, Z. Molecular Phylogeny and Adaptive Mitochondrial DNA Evolution of Salmonids (Pisces: Salmonidae). Front. Genet. 2022, 13, 903240. [Google Scholar] [CrossRef]
- Nelson, J.; Grande, T.C.; Wilson, M.V. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Cai, J.; Zhou, X.; Yan, X.; Lucente, D.; Lagana, C. Top 10 Species Groups in Global Aquaculture 2017. Food and Agriculture Organization of the United Nations, FAO Fisheries and Aquaculture Department. 2019. Available online: http://www.fao.org/3/ca5224en/CA5224EN.pdf (accessed on 12 October 2022).
- FAO. The State of World Fisheries and Aquaculture 2020. In Sustainability in Action; FAO: Rome, Italy, 2020; Volume 32, p. 244. [Google Scholar] [CrossRef]
- Phillips, R.B.; Oakley, T.H. Phylogenetic relationships among the Salmonidae based on nuclear DNA and mitochondrial DNA sequences. In Molecular Systematics of Fishes; Kocher, T., Stepien, C., Eds.; Academic Press: San Diego, CA, USA, 1997; pp. 145–162. [Google Scholar]
- Wilhelm, V.; Villegas, J.; Miquel, A.; Engel, E.; Bernales, S.; Valenzuela, P.D.; Burzio, L.O. The complete sequence of the mitochondrial genome of the Chinook salmon, Oncorhynchus tshawytscha. Biol. Res. 2003, 36, 223–231. [Google Scholar] [CrossRef]
- Suckley, G. Notices of certain New Species of North American Salmonidæ, chiefly in the Collection of the NW Boundary Commission, in charge of Archibald Campbell, Esq., Commissioner of the United States, collected by Doctor CBR Kennerly, Naturalist to the Commission. Ann. Lyceum Nat. Hist. N. Y. 1862, 7, 306–313. [Google Scholar] [CrossRef]
- Hendrickson, D.A.; Perez, H.E.; Findley, L.T.; Forbes, W.; Tomelleri, J.R.; Mayden, R.L.; Nielsen, J.L.; Jensen, B.; Campos, G.R.; Romero, A.V.; et al. Mexican native trouts: A review of their history and current systematic and conservation status. Rev. Fish Biol. Fish. 2002, 12, 273–316. [Google Scholar] [CrossRef]
- Espinosa-Pérez, H.; García-De León, F.J.; Ruiz-Campos, G.; Varela-Romero, A.; Barriga-Sosa, I.D.L.A.; Arredondo-Figueroa, J.; Hendrickson, D.A.; Camarena-Rosales, F.; De los Santos-Camarillo, A.B. Las Truchas Mexicanas. Especies 2007, 16, 9–14. [Google Scholar]
- Mayden, R.L.; Dillman, C.B.; Espinosa-Pérez, H.; Tomelleri, J.R.; Kuhajda, B.R.; Hendrickson, D.A.; Ruiz-Campos, G.; De los Santos-Camarillo, A.B.; García-De León, F.J.; Varela-Romero, A.; et al. Evolution and Diversity of Trout Species in México. In Conserving Wild Trout, Proceedings of the Wild Trout X Symposium, Bozeman, Montana, 28–29 September 2010; Carline, R.F., Lo Sapio, C., Eds.; USDA Forest Service, Ecosystem Management Coordination, WOD, Publishing Arts, Carol LoSapio: West Yellowstone, MT, USA, 2010; pp. 134–144. [Google Scholar]
- Needham, P.R.; Gard, R. A New Trout from Central Mexico: Salmo chrysogaster, the Mexican Golden Trout. Copeia 1964, 1964, 169–173. [Google Scholar] [CrossRef]
- Evermann, B.W. Descriptions of a new species of trout (Salmo nelsoni) and a new cyprinodont (Fundulus meeki) with notes on other fishes from Lower California. Proc. Biol. Soc. Wash. 1908, 21, 19–30. [Google Scholar]
- Ruiz-Campos, G. Bionomía y Ecología Poblacional de la Trucha Arcoíris, Onchorhynchus mykiss Nelsoni (Everman), de la Sierra San Pedro Mártir, Baja California, México. Ph.D. Thesis, Universidad de Nuevo León, Monterrey, Mexico, 1993. [Google Scholar]
- Ruiz-Campos, G.; Pister, E.P. Distribution, habitat, and current status of the San Pedro Martir Rainbow trout, Oncorhynchus mykiss nelsoni. Bull. South Calif. Acad. Sci. 1995, 94, 131–148. [Google Scholar]
- FAO. Oncorhynchus mykiss. In Cultured Aquatic Species Fact Sheets; Crespi, V., New, M., Eds.; Multilingual, Text by Cowx, I.G., CD-ROM; FAO: Rome, Italy, 2009. [Google Scholar]
- Abadía-Cardoso, A.; Pearse, D.E.; Jacobson, S.; Marshall, J.; Dalrymple, D.; Kawasaki, F.; Ruiz-Campos, G.; Garza, J.C. Population genetic structure and ancestry of steelhead/rainbow trout (Oncorhynchus mykiss) at the extreme southern edge of their range in North America. Conserv. Genet. 2016, 17, 675–689. [Google Scholar] [CrossRef]
- Behnke, R.J. Trout and Salmon of North America; Chanticleer Press, Inc.: New York, NY, USA, 2002. [Google Scholar]
- Hendrickson, D.A.; Neely, D.A.; Mayden, R.L.; Anderson, K.; Brooks, J.E.; Camerana-Rosales, F.; Cutter, R.F.; Cutter, L.; De los Santos-Camarillo, A.B.; Ernsting, G.W.; et al. Conservation of Mexican native trout and the discovery, status, protection and recovery of the Conchos trout, the first native Oncorhynchus of the Atlantic drainage in Mexico. In UT Faculty/Researcher Works; Dirección de Publicaciones, Universidad Autónoma de Nuevo León, Facultad de Ciencias Biológicas: Monterrey, Mexico, 2007. [Google Scholar]
- Mayden, R.L. Biodiversity of Mexican trout (Teleostei: Salmonidae: Oncorhynchus): Recent findings conservation concerns, and management recommendations. In Homenaje al Doctor Andrés Reséndez-Medina; Lozano-Vilano, M.L., Contreras-Balderas, A.J., Eds.; Universidad Autónoma de Nuevo León: Monterrey, Mexico, 2004; pp. 269–282. [Google Scholar]
- Camarena-Rosales, F.; Ruiz-Campos, G.; De La Rosa-Vélez, J.; Mayden, R.L.; Hendrickson, D.A.; Varela-Romero, A.; de León, F.J.G. Mitochondrial haplotype variation in wild trout populations (Teleostei: Salmonidae) from northwestern Mexico. Rev. Fish Biol. Fish. 2007, 18, 33–45. [Google Scholar] [CrossRef]
- Varela-Romero, A.; Hendrickson, D.A. Los peces dulceacuícolas de Sonora. In Diversidad Biológica de Sonora; Molina-Fraener, F., Van Devender, T., Eds.; UNAM: Mexico City, Mexico, 2009. [Google Scholar]
- Escalante, M.A.; García-De-León, F.J.; Dillman, C.B.; Camarillo, A.D.L.S.; George, A.; Barriga-Sosa, I.D.L.A.; Ruiz-Luna, A.; Mayden, R.L.; Manel, S. Genetic introgression of cultured rainbow trout in the Mexican native trout complex. Conserv. Genet. 2014, 15, 1063–1071. [Google Scholar] [CrossRef]
- Escalante, M.A.; León, F.J.G.; Ruiz-Luna, A.; Landguth, E.; Manel, S. The interplay of riverscape features and exotic introgression on the genetic structure of the Mexican golden trout (Oncorhynchus chrysogaster), a simulation approach. J. Biogeogr. 2018, 45, 1500–1514. [Google Scholar] [CrossRef]
- Escalante, M.A.; Perrier, C.; León, F.J.G.-D.; Ruiz-Luna, A.; Ortega-Abboud, E.; Manel, S. Genotyping-by-sequencing reveals the effects of riverscape, climate and interspecific introgression on the genetic diversity and local adaptation of the endangered Mexican golden trout (Oncorhynchus chrysogaster). Conserv. Genet. 2020, 21, 907–926. [Google Scholar] [CrossRef]
- Abadía-Cardoso, A.; Garza, J.C.; Mayden, R.L.; de León, F.J.G. Genetic Structure of Pacific Trout at the Extreme Southern End of Their Native Range. PLoS ONE 2015, 10, e0141775. [Google Scholar] [CrossRef] [PubMed]
- Abadía-Cardoso, A.; García-De León, F.J.; Garza, J.C. Historia evolutiva y biodiversidad genética de las truchas de la Sierra Madre Occidental. In La Trucha Dorada Mexicana; Ruiz-Luna, A., García-De León, F.J., Eds.; CIAD, CIB, CONACYT, La Paz, BCS; Eddel Graph, S.A. de C.V.: Ciudad de Mexico, Mexico, 2016; pp. 29–38. [Google Scholar]
- Ballesteros-Córdova, C.A.; Varela-Romero, A.V.; Grijalva-Chon, J.M.; Castillo-Gámez, R.A.; Camarena-Rosales, F.; Ruiz-Campos, G. Variabilidad genética poblacional de la trucha Yaqui (Oncorhynchus sp.) en la región de Mesa Tres Ríos, Sonora, México. Biotecnia 2019, 21, 134–142. [Google Scholar] [CrossRef]
- León, F.J.G.-D.; Dillman, C.B.; Camarillo, A.B.D.L.S.; George, A.L.; Camarena-Rosales, F.; Barriga-Sosa, I.D.L.A.; Mayden, R.L. First steps towards the identification of evolutionarily significant units in Mexican native trout: An assessment of microsatellite variation. Environ. Biol. Fishes 2020, 103, 733–756. [Google Scholar] [CrossRef]
- Lecaudey, L.A.; Schliewen, U.K.; Osinov, A.G.; Taylor, E.B.; Bernatchez, L.; Weiss, S.J. Inferring phylogenetic structure, hybridization and divergence times within Salmoninae (Teleostei: Salmonidae) using RAD-sequencing. Mol. Phylogenetics Evol. 2018, 124, 82–99. [Google Scholar] [CrossRef] [PubMed]
- Shedko, S.V.; Miroshnichenko, I.L.; Nemkova, G.A. Phylogeny of salmonids (salmoniformes: Salmonidae) and its molecular dating: Analysis of mtDNA data. Russ. J. Genet. 2013, 49, 623–637. [Google Scholar] [CrossRef]
- Shedko, S.V.; Miroshnichenko, I.L.; Nemkova, G.A. Phylogeny of salmonids (Salmoniformes: Salmonidae) and its molecular dating: Analysis of nuclear RAG1 gene. Russ. J. Genet. 2012, 48, 575–579. [Google Scholar] [CrossRef]
- Crête-Lafrenière, A.; Weir, L.K.; Bernatchez, L. Framing the Salmonidae Family Phylogenetic Portrait: A More Complete Picture from Increased Taxon Sampling. PLoS ONE 2012, 7, e46662. [Google Scholar] [CrossRef]
- Behnke, R.J. Native Trout of Western North America. In American Fisheries Society Monograph 6; American Fisheries Society: Bethesda, MD, USA, 1992. [Google Scholar]
- Crespi, B.J.; Fulton, M.J. Molecular systematics of Salmonidae: Combined nuclear data yields a robust phylogeny. Mol. Phylogenet. Evol. 2004, 31, 658–679. [Google Scholar] [CrossRef]
- Loudenslager, E.J.; Rinne, J.N.; Gall, G.A.E.; David, R.E. Biochemical Genetic Studies of Native Arizona and New Mexico Trout. Southwest. Nat. 1986, 31, 221–234. [Google Scholar] [CrossRef]
- Nielsen, J.L.; Fountain, M.C.; Wright, J.M. Biogeographic analysis of Pacific trout (Oncorhynchus mykiss) in California and Mexico based on mtDNA and nuclear microsatellites. In Molecular Systematics of Fishes; Kocher, T., Stepien, C.A., Eds.; Academic Press: San Diego, CA, USA, 1997; pp. 53–73. [Google Scholar]
- Nielsen, J.L.; Fountain, M.C.; Favela, J.C.; Cobble, K.; Jensen, B.L. Oncorhynchus at the southern extent of their range: A study of mtDNA control–region sequence with special reference to an undescribed subspecies of O. mykiss from Mexico. Environ. Biol. Fishes 1998, 51, 7–23. [Google Scholar] [CrossRef]
- Glenn, T.C.; Nilsen, R.A.; Kieran, T.; Sanders, J.G.; Bayona-Vásquez, N.J.; Finger, J.; Pierson, T.W.; Bentley, K.E.; Hoffberg, S.L.; Louha, S.; et al. Adapterama I: Universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext). PeerJ 2019, 7, e7755. [Google Scholar] [CrossRef]
- Rohland, N.; Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012, 22, 939–946. [Google Scholar] [CrossRef] [PubMed]
- National Library of Medicine. National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov (accessed on 13 February 2022).
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Baeza, J.A. An introduction to the Special Section on Crustacean Mitochondrial Genomics: Improving the assembly, annotation, and characterization of mitochondrial genomes using user-friendly and open-access bioinformatics tools, with decapod crustaceans as an example. J. Crustac. Biol. 2022, 42, ruac012. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics 2008, 24, 861–862. [Google Scholar] [CrossRef]
- Stothard, P.; Hiseni, P.; Wilson, R.C.; Storrø, O.; Johnsen, R.; Øien, T.; Rudi, K. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Cucini, C.; Leo, C.; Iannotti, N.; Boschi, S.; Brunetti, C.; Pons, J.; Fanciulli, P.P.; Frati, F.; Carapelli, A.; Nardi, F. EZmito: A simple and fast tool for multiple mitogenome analyses. Mitochondrial DNA Part B 2021, 6, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-P.; Wan, H.-L.; Zhang, S.; Yu, J. γ-MYN: A new algorithm for estimating Ka and Ks with consideration of variable substitution rates. Biol. Direct 2009, 4, 20. [Google Scholar] [CrossRef]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2011, 40, 2833–2845. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4.4. [Place unknown]: [Publisher unknown]. 2018. Available online: https://github.com/rambaut/figtree/releases/tag/v1.4.4 (accessed on 22 March 2023).
- Wang, F.; He, E.; Li, Y.; Cai, X.; Ma, W. Complete mitochondrial genome of the hybridized fish (Oncorhynchus mykiss ♀ × Atlantic salmon ♂). Mitochondrial DNA Part A 2015, 27, 4153–4154. [Google Scholar] [CrossRef] [PubMed]
- Schroeter, J.C.; Maloy, A.P.; Rees, C.B.; Bartron, M.L. Fish mitochondrial genome sequencing: Expanding genetic resources to support species detection and biodiversity monitoring using environmental DNA. Conserv. Genet. Resour. 2019, 12, 433–446. [Google Scholar] [CrossRef]
- Si, S.; Wang, Y.; Xu, G.; Yang, S.; Mou, Z.; Song, Z. Complete mitochondrial genomes of two lenoks, Brachymystax lenok and Brachymystax lenok tsinlingensis. Mitochondrial DNA 2012, 23, 338–340. [Google Scholar] [CrossRef]
- Barriga-Sosa, I.D.L.A.; De León, F.J.G.; Del Río-Portilla, M.A. The complete mitochondrial DNA of the endemic shortfin silverside, Chirostoma humboldtianum (Valenciennes, 1835). Mitochondrial DNA 2014, 27, 1545–1546. [Google Scholar] [CrossRef]
- Galván-Tirado, C.; del Río-Portilla, M.A.; Delgado-Vega, R.; León, F.J.G.-D. Genetic variability between complete mitochondrion genomes of the sablefish, Anoplopoma fimbria (Pallas, 1814). Mitochondrial DNA Part A 2015, 27, 2429–2430. [Google Scholar] [CrossRef]
- Magallón-Gayón, E.; del Río-Portilla, M.; Barriga-Sosa, I.D.L.A. The complete mitochondrial genomes of two octopods of the eastern Pacific Ocean: Octopus mimus and ‘Octopus’ fitchi (Cephalopoda: Octopodidae) and their phylogenetic position within Octopoda. Mol. Biol. Rep. 2019, 47, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Machkour-M’Rabet, S.; Hanes, M.M.; Martínez-Noguez, J.J.; Cruz-Medina, J.; León, F.J.G.-D. The queen conch mitogenome: Intra- and interspecific mitogenomic variability in Strombidae and phylogenetic considerations within the Hypsogastropoda. Sci. Rep. 2021, 11, 11972. [Google Scholar] [CrossRef]
- Colín, A.; Galván-Tirado, C.; Carreón-Palau, L.; Bracken-Grissom, H.D.; Baeza, J.A. Mitochondrial genomes of the land hermit crab Coenobita clypeatus (Anomura: Paguroidea) and the mole crab Emerita talpoida (Anomura: Hippoidea) with insights into phylogenetic relationships in the Anomura (Crustacea: Decapoda). Gene 2023, 849, 146896. [Google Scholar] [CrossRef]
- Zardoya, R.; Garrido-Pertierra, A. The complete nucleotide sequence of the mitochondrial DNA genome of the rainbow trout, Oncorhynchus mykiss. J. Mol. Evol. 1995, 41, 942–951. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Yang, S.; Song, Z. The complete mitochondrial genome of the taimen, Hucho taimen, and its unusual features in the control region. Mitochondrial DNA 2011, 22, 111–119. [Google Scholar] [CrossRef]
- Yu, J.-N.; Kwak, M. The complete mitochondrial genome of Brachymystax lenok tsinlingensis (Salmoninae, Salmonidae) and its intraspecific variation. Gene 2015, 573, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-G.; Li, Y.-Y.; Meng, W.; Liu, L.-X. The complete mitochondrial DNA sequence of Xinjiang arctic grayling Thymallus arcticus grubei. Mitochondrial DNA Part B 2016, 1, 724–725. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Meng, F.; Wang, R.; Shi, G. Complete mitochondrial genome of the Salvelinus malma sp. (Salmoniformes, Salmonidae) with phylogenetic consideration. Mitochondrial DNA Part B 2017, 2, 889–890. [Google Scholar] [CrossRef]
- Miya, M.; Takeshima, H.; Endo, H.; Ishiguro, N.B.; Inoue, J.G.; Mukai, T.; Satoh, T.P.; Yamaguchi, M.; Kawaguchi, A.; Mabuchi, K.; et al. Major patterns of higher teleostean phylogenies: A new perspective based on 100 complete mitochondrial DNA sequences. Mol. Phylogenetics Evol. 2003, 26, 121–138. [Google Scholar] [CrossRef]
- Shirai, K.; Inomata, N.; Mizoiri, S.; Aibara, M.; Terai, Y.; Okada, N.; Tachida, H. High prevalence of non-synonymous substitutions in mtDNA of cichlid fishes from Lake Victoria. Gene 2014, 552, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Zhou, L.; Yang, W.-T.; Miao, L.-J.; Li, Z.; Zhang, X.-J.; Wang, Y.; Gui, J.-F. Comparative mitogenome analyses uncover mitogenome features and phylogenetic implications of the subfamily Cobitinae. BMC Genom. 2021, 22, 50. [Google Scholar] [CrossRef]
- Castellana, S.; Vicario, S.; Saccone, C. Evolutionary Patterns of the Mitochondrial Genome in Metazoa: Exploring the Role of Mutation and Selection in Mitochondrial Protein–Coding Genes. Genome Biol. Evol. 2011, 3, 1067–1079. [Google Scholar] [CrossRef]
- Johansen, S.; Guddal, P.H.; Johansen, T. Organization of the mitochondrial genome of Atlantic cod, Gadus morhua. Nucleic Acids Res. 1990, 18, 411–419. [Google Scholar] [CrossRef]
- Rinne, J.N.; Minckley, W.L. Patterns of Variation and Distribution in Apache Trout (Salmo apache) Relative to Co-Occurrence with Introduced Salmonids. Copeia 1985, 1985, 285–292. [Google Scholar] [CrossRef]
- Jordan, D.S. Description of a new subspecies of Trout from McCloud River, California. Proc. Acad. Nat. Sci. Phila. 1894, XLVI, 60. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).