Costimulatory Molecules CD80/86 Trigger Non-Specific Cytotoxic Cell of Nile tilapia (Oreochromis niloticus) to Kill CIK Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Preparing and Sample Collection
2.2. Construction, Expression and Purification of the Recombinant On-CD80/86 (rOn-CD80/86) Plasmid
2.3. Western Blot Analysis
2.4. Activation and Regulation of rOn-CD80/86 on NCC Activity
2.5. Assay for the Killing Effect of NCCs
2.6. Detection of Interaction between On-CD80/86 and On-NCCRP-1
2.7. Expression Analysis of On-CD28 and On-NCCRP-1 in the Head Kidney Leukocytes (HKLs)
2.8. Statistical Analysis
3. Results
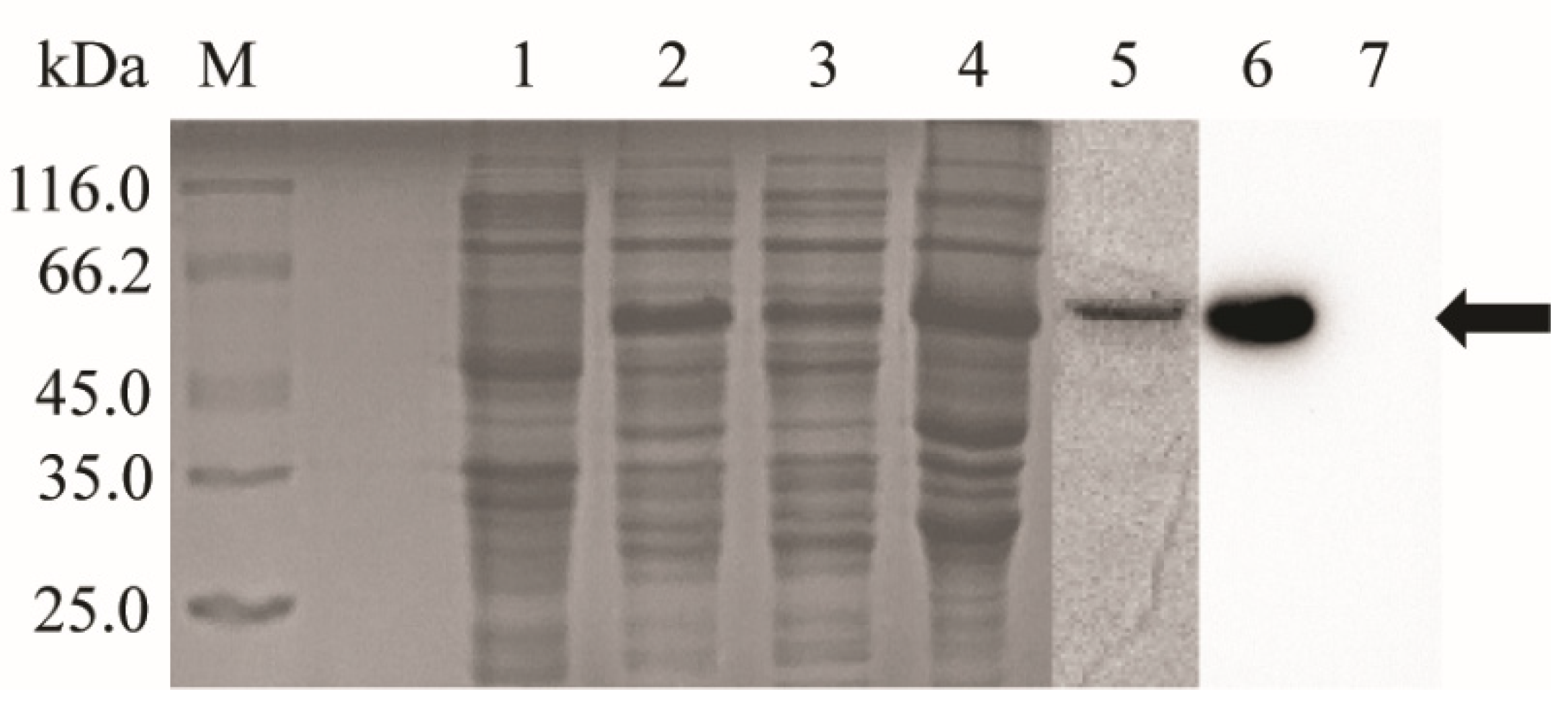
3.1. Recombinant On-CD80/86 Expression, Purification and Western Blot Analysis
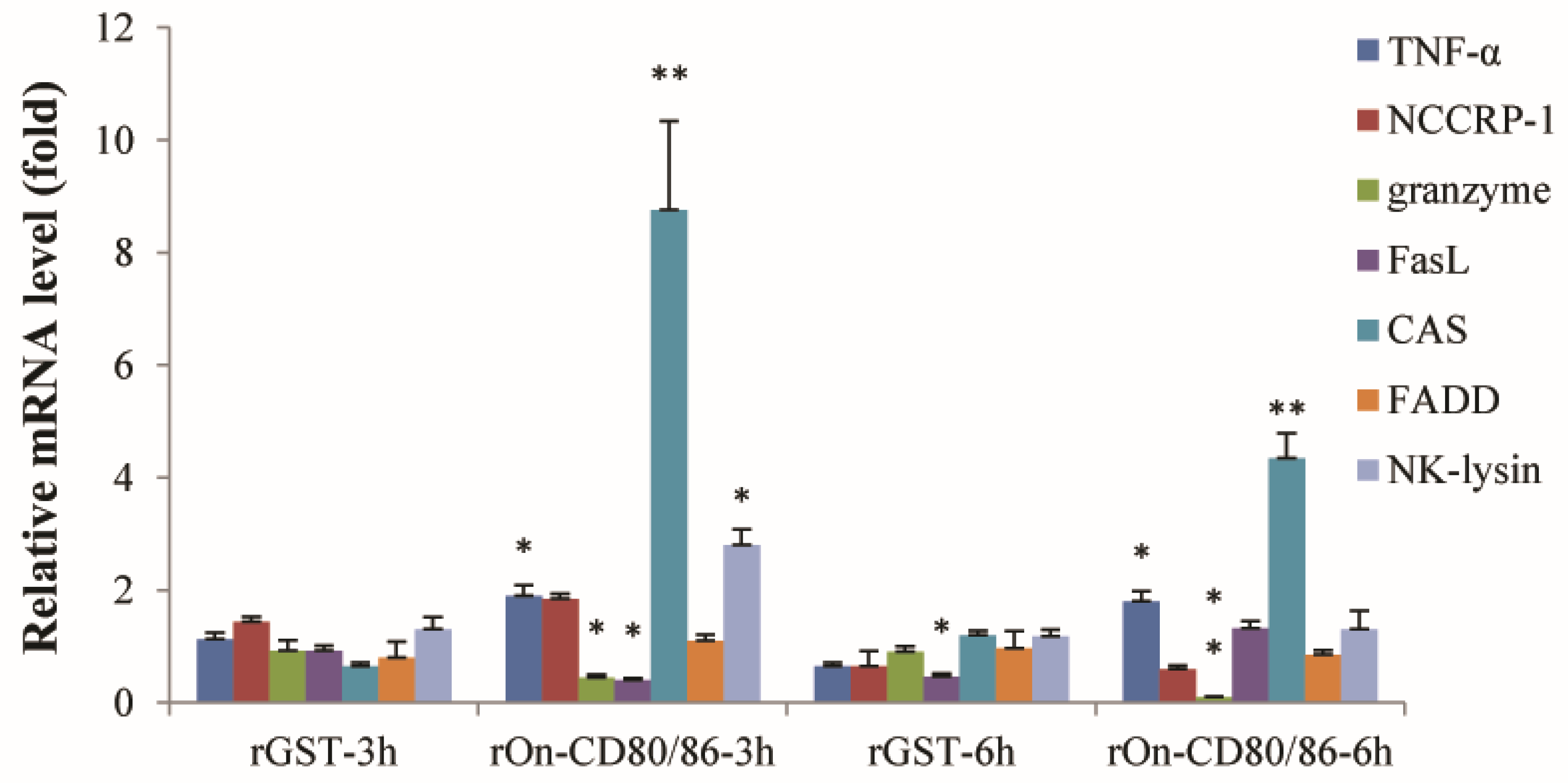
3.2. Effect of rOn-CD80/86 Protein on the Expression of NCC Effector Molecules
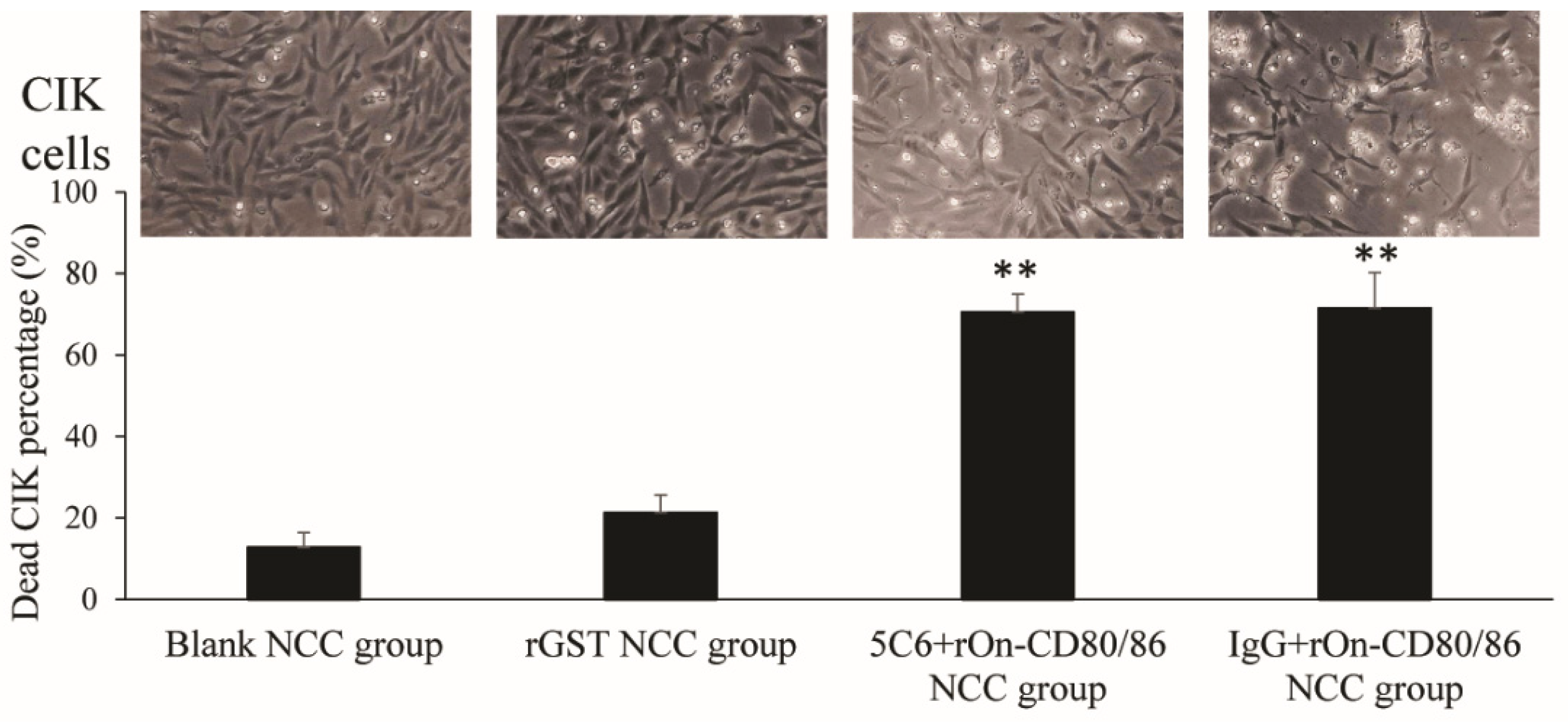
3.3. Effect of rOn-CD80/86 Protein on the Cytotoxicity of NCC
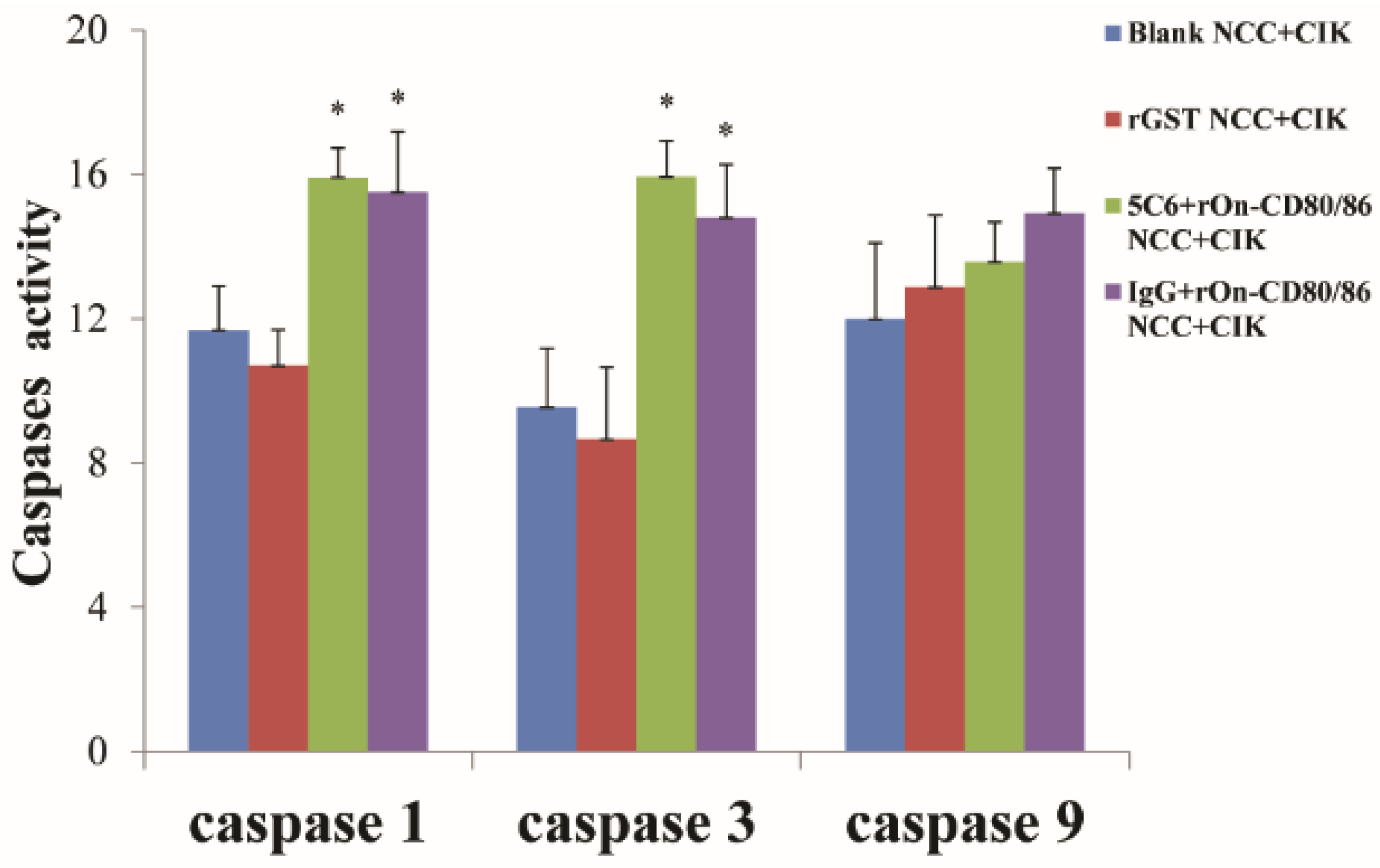
3.4. Activity of Caspase-1, 3, and 9 on CIK
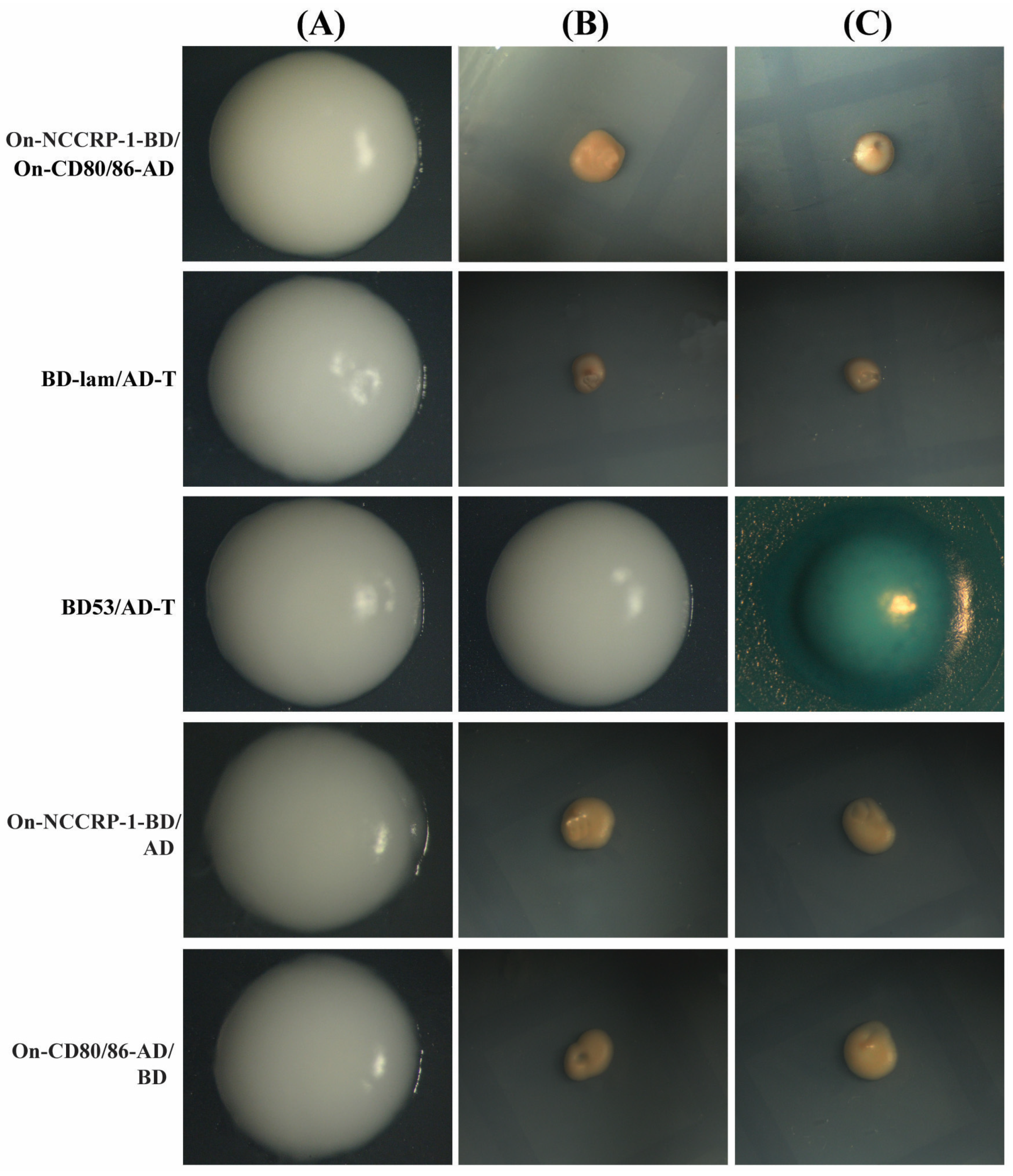
3.5. Interaction of On-NCCRP-1 with On-CD80/86
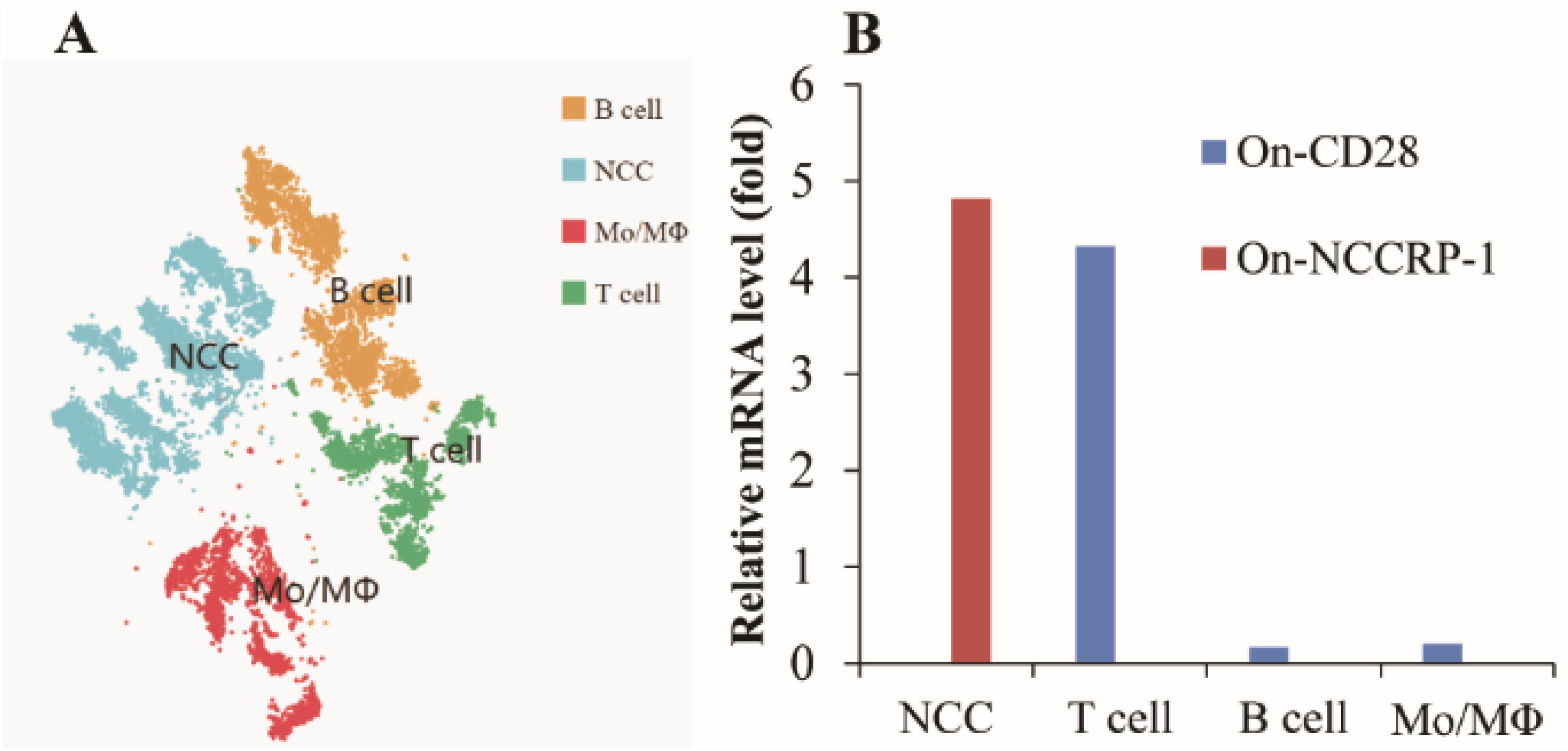
3.6. Expression Characteristics of On-CD28 and On-NCCRP-1 Genes in Nile tilapia HKLs scRNA-Seq
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaso-Friedmann, L.; Leary, J.H., 3rd; Evans, D.L. The non-specific cytotoxic cell receptor (NCCRP-1): Molecular organization and signaling properties. Dev. Comp. Immunol. 2001, 25, 701–711. [Google Scholar] [CrossRef]
- Praveen, K.; Leary, J.H., 3rd; Evans, D.L.; Jaso-Friedmann, L. Nonspecific cytotoxic cells of teleosts are armed with multiple granzymes and other components of the granule exocytosis pathway. Mol. Immunol. 2006, 43, 1152–1162. [Google Scholar] [CrossRef]
- Evans, D.L.; Leary, J.H., 3rd; Jaso-Friedmann, L. Nonspecific cytotoxic cells and innate immunity: Regulation by programmed cell death. Dev. Comp. Immunol. 2001, 25, 791–805. [Google Scholar] [CrossRef]
- Cuesta, A.; Esteban, M.; Meseguer, J. Molecular characterization of the nonspecific cytotoxic cell receptor (NCCRP-1) demonstrates gilthead seabream NCC heterogeneity. Dev. Comp. Immunol. 2005, 29, 637–650. [Google Scholar] [CrossRef]
- Greenlee, A.R.; Brown, R.A.; Ristow, S.S. Nonspecific cytotoxic cells of rainbow trout (Oncorhynchus mykiss) kill YAC-1 targets by both necrotic and apoptic mechanisms. Dev. Comp. Immunol. 1991, 15, 153–164. [Google Scholar] [CrossRef]
- Jaso-Friedmann, L.; Leary, J.H., 3rd; Evans, D.L. Role of nonspecific cytotoxic cells in the induction of programmed cell death of pathogenic protozoans: Participation of the Fas ligand-Fas receptor system. Exp. Parasitol. 2000, 96, 75–88. [Google Scholar] [CrossRef]
- Praveen, K.; Evans, D.L.; Jaso-Friedmann, L. Constitutive expression of tumor necrosis factor-alpha in cytotoxic cells of teleosts and its role in regulation of cell-mediated cytotoxicity. Mol. Immunol. 2006, 43, 279–291. [Google Scholar] [CrossRef]
- Meseguer, J.; Esteban, M.A.; Mulero, V. Nonspecific cell-mediated cytotoxicity in the seawater teleosts (Sparus aurata and Dicentrarchus labrax): Ultrastructural study of target cell death mechanisms. Anat. Rec. 1996, 244, 499–505. [Google Scholar] [CrossRef]
- Reimers, K.; Abu Qarn, M.; Allmeling, C.; Bucan, V.; Vogt, P.M. Identification of the non-specific cytotoxic cell receptor protein 1 (NCCRP1) in regenerating axolotl limbs. J. Comp. Physiol. B 2006, 176, 599–605. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Sebzda, E.; Kündig, T.M.; Shahinian, A.; Speiser, D.; Mak, T.W.; Ohashi, P.S. T cell responses are governed by avidity and co-stimulatory thresholds. Eur. J. Immunol. 1996, 26, 2017–2022. [Google Scholar] [CrossRef]
- Boomer, J.S.; Green, J.M. An enigmatic tail of CD28 signaling. Cold Spring Harb. Perspect. Biol. 2010, 2, a002436. [Google Scholar] [CrossRef]
- Martín-Fontecha, A.; Assarsson, E.; Carbone, E.; Kärre, K.; Ljunggren, H.G. Triggering of murine NK cells by CD40 and CD86 (B7-2). J. Immunol. 1999, 162, 5910–5916. [Google Scholar]
- Wilson, J.L.; Charo, J.; Martín-Fontecha, A.; Dellabona, P.; Casorati, G.; Chambers, B.J.; Kiessling, R.; Bejarano, M.T.; Ljunggren, H.G. NK cell triggering by the human costimulatory molecules CD80 and CD. J. Immunol. 1999, 163, 4207–4212. [Google Scholar]
- Zhang, X.J.; Zhang, X.Y.; Wang, P.; Zhang, Y.A. Identification of another primordial CD80/86 molecule in rainbow trout: Insights into the origin and evolution of CD80 and CD86 in vertebrates. Dev. Comp. Immunol. 2018, 89, 73–82. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Z.; Zheng, Q.; Tang, J.; Cai, J.; Lu, Y.; Jian, J. Conservation of structural and interactional features of CD28 and CD80/86 molecules from Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2018, 72, 95–103. [Google Scholar] [CrossRef]
- Lu, T.Z.; Liu, X.; Wu, C.S.; Ma, Z.Y.; Wang, Y.; Zhang, Y.A.; Zhang, X.J. Molecular and Functional Analyses of the Primordial Costimulatory Molecule CD80/86 and Its Receptors CD28 and CD152 (CTLA-4) in a Teleost Fish. Front. Immunol. 2022, 13, 885005. [Google Scholar] [CrossRef]
- Teng, J.; Cui, M.Y.; Zhao, Y.; Chen, H.J.; Du, W.J.; Xue, L.Y.; Ji, X.S. Expression changes of non-specific cytotoxic cell receptor (NCCRP1) and proliferation and migration of NCCs post-Nocardia seriolae infection in Northern Snakehead. Dev. Comp. Immunol. 2022, 139, 104576. [Google Scholar] [CrossRef]
- Huang, X.Z.; Li, Y.W.; Mai, Y.Z.; Luo, X.C.; Dan, X.M.; Li, A.X. Molecular cloning of NCCRP-1 gene from orange-spotted grouper (Epinephelus coioides) and characterization of NCCRP-1(+) cells post Cryptocaryon irritans infection. Dev. Comp. Immunol. 2014, 46, 267–278. [Google Scholar] [CrossRef]
- Zhao, X.L.; Han, Y.; Ren, S.T.; Ma, Y.M.; Li, H.; Peng, X.X. L-proline increases survival of tilapias infected by Streptococcus agalactiae in higher water temperature. Fish Shellfish Immunol. 2015, 44, 33–42. [Google Scholar] [CrossRef]
- Maulu, S.; Hasimuna, O.J.; Mphande, J.; Munang’Andu, H.M. Prevention and Control of Streptococcosis in Tilapia Culture: A Systematic Review. J. Aquat. Anim. Health 2021, 33, 162–177. [Google Scholar] [CrossRef]
- Li, Q.; Jiang, B.; Zhang, Z.; Huang, Y.; Xu, Z.; Chen, X.; Huang, Y.; Jian, J. SP protects Nile tilapia (Oreochromis niloticus) against acute Streptococcus agalatiae infection. Fish Shellfish Immunol. 2022, 123, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zheng, Q.; Niu, J.; Tang, J.; Wang, B.; Abarike, E.D.; Lu, Y.; Cai, J.; Jian, J. NK-lysin from Oreochromis niloticus improves antimicrobial defence against bacterial pathogens. Fish Shellfish Immunol. 2018, 72, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zheng, Q.; Wang, Z.; Tang, J.; Lu, Y.; Qin, Q.; Cai, J.; Jian, J. Fish natural killer enhancing factor-A (NKEF-A) enhance cytotoxicity of nonspecific cytotoxic cells against bacterial infection. Mol. Immunol. 2021, 133, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, Z.; Xie, R.; Wang, P.; Zhang, Z.; Cai, J.; Wang, B.; Jian, J. Transferrin Mediated NCC Killing Activity through NCCRP-1 in Nile tilapia (Oreochromis niloticus). Fishes 2022, 7, 253. [Google Scholar] [CrossRef]
- Niu, J.; Huang, Y.; Liu, X.; Zhang, Z.; Tang, J.; Wang, B.; Lu, Y.; Cai, J.; Jian, J. Single-cell RNA-seq reveals different subsets of non-specific cytotoxic cells in teleost. Genomics 2020, 112, 5170–5179. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, Y.; Savan, R.; Endo, M.; Sakai, M.; Immunology, J.F. Non-specific cytotoxic cell receptor (NCCRP)-1 type gene in tilapia (Oreochromis niloticus): Its cloning and analysis. Fish Shellfish Immunol. 2004, 16, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Parida, S.; Mohapatra, A.; Sahoo, P.K. Cloning and functional characterisation of natural killer enhancing factor-B (NKEF-B) gene of Labeo rohita: Anti-oxidant and antimicrobial activities of its recombinant protein. Mol. Immunol. 2020, 126, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Bots, M.; Medema, J.P. Granzymes at a glance. J. Cell Sci. 2006, 119, 5011–5014. [Google Scholar] [CrossRef] [PubMed]
- Walch, M.; Dotiwala, F.; Mulik, S.; Thiery, J.; Kirchhausen, T.; Clayberger, C.; Krensky, A.M.; Martinvalet, D.; Lieberman, J. Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell 2014, 157, 1309–1323. [Google Scholar] [CrossRef] [PubMed]
- Praveen, K.; Leary, J.H.; Evans, D.L.; Jaso-Friedmann, L. Molecular cloning of cellular apoptosis susceptibility (CAS) gene in Oreochromis niloticus and its proposed role in regulation of non-specific cytotoxic cell (NCC) functions. Fish Shellfish Immunol. 2006, 20, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Gunne, H.; Agerberth, B.; Boman, A.; Bergman, T.; Olsson, B.; Dagerlind, A.; Wigzell, H.; Boman, H.; Gudmundsson, G. NK-lysin, structure and function of a novel effector molecule of porcine T and NK cells. Vet. Immunol. Immunopathol. 1996, 54, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Liu, Y.P.; Tong, X.X.; Zhang, T.N.; Yang, N. Molecular mechanisms and functions of pyroptosis in sepsis and sepsis-associated organ dysfunction. Front. Cell. Infect. Microbiol. 2022, 12, 962139. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, N.; Collignon, T.E.; Tewari, D.; Bishayee, A. Hypericin and its anticancer effects: From mechanism of action to potential therapeutic application. Phytomedicine 2022, 105, 154356. [Google Scholar] [CrossRef] [PubMed]
- Bluestone, J.A. New perspectives of CD28-B7-mediated T cell costimulation. Immunity 1995, 2, 555–559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Paiano, A.; Margiotta, A.; De Luca, M.; Bucci, C. Yeast Two-Hybrid Assay to Identify Interacting Proteins. Curr. Protoc. Protein Sci. 2019, 95, e70. [Google Scholar] [CrossRef] [PubMed]
| Primers | Sequences (5′→3′) | Purpose |
|---|---|---|
| point-to-point Y2H | On-NCCRP-1-S | GGAATTCATGTCTGCTGCCGAGTGGAAG |
| point-to-point Y2H | On-NCCRP-1-A | GGGATCCCGCGGGCTGCTTTTGCTTGGTC |
| point-to-point Y2H | pGADT7-On-CD80/86-S | CGGGATCCATACAGGAAGTGCTAAATTTCTC |
| point-to-point Y2H | pGADT7-On-CD80/86-A | CCGCTCGAGCTAATCAGTGCTGTCTTGTATCTG |
| Protein expression | CD80/86-EcoRI-F | TCCGAATTCTTCACCGTTACCGGTTCTGC |
| Protein expression | CD80/86-XhoI-R | TGCTCGAGAGAACCACCGGTCTGACGACG |
| qRT-PCR | qβ-actin-S | AGATGAAATCGCCGCACTGG |
| qRT-PCR | qβ-actin-A | TCTGACCCATACCCACCATCA |
| qRT-PCR | RT-TNFα-F | CTCGTCGTCGTGGCTCTTT |
| qRT-PCR | RT-TNFα-R | CCTTGGCTTTGCTGCTGAT |
| qRT-PCR | RT-NCCRP1-F | CACCACCTGAACCCGAACT |
| qRT-PCR | RT-NCCRP1-R | GGTCCACAACCTGCTCCAT |
| qRT-PCR | RT-granzyme-F | ATACAACTGGCAAGGAAGGAG |
| qRT-PCR | RT-granzyme-R | TACCCATCTCAGCACATCAAC |
| qRT-PCR | RT-FasL-F | CTTCTCCAAGGGCGATTCTA |
| qRT-PCR | RT-FasL-R | ATCTCCCTGAGTGGCTGTGC |
| qRT-PCR | RT-CAS-F | CAGCAGTTTCGAGGAAGCAC |
| qRT-PCR | RT-CAS-R | TCCAAGCAAGCCAGGTATTT |
| qRT-PCR | RT-FADD-F | ACTGGCAGAAGATAACACGG |
| qRT-PCR | RT-FADD-R | TTTGCTTTCTCCTCCTCACT |
| qRT-PCR | RT-NK-lysin-F | ATTTGCGGCACAGTGATTT |
| qRT-PCR | RT-NK-lysin-R | ATGGAAGTCTTGATGGGGCT |
| Protein | Accession No. |
|---|---|
| CD80/86 | MF150103.1 |
| TNFα | AY428948.1 |
| NCCRP-1 | MF162296 |
| Granzyme | AY918866.1 |
| FasL | KM008610.1 |
| CAS | AF547173 |
| FADD | XM_003456561.5 |
| NK-lysin | ATW66454.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Chen, Z.; Xie, R.; Amoah, K.; Wang, B.; Cai, J.; Lu, Y.; Jian, J. Costimulatory Molecules CD80/86 Trigger Non-Specific Cytotoxic Cell of Nile tilapia (Oreochromis niloticus) to Kill CIK Cells. Fishes 2022, 7, 353. https://doi.org/10.3390/fishes7060353
Huang Y, Chen Z, Xie R, Amoah K, Wang B, Cai J, Lu Y, Jian J. Costimulatory Molecules CD80/86 Trigger Non-Specific Cytotoxic Cell of Nile tilapia (Oreochromis niloticus) to Kill CIK Cells. Fishes. 2022; 7(6):353. https://doi.org/10.3390/fishes7060353
Chicago/Turabian StyleHuang, Yu, Zhengsi Chen, Ruitao Xie, Kwaku Amoah, Bei Wang, Jia Cai, Yishan Lu, and Jichang Jian. 2022. "Costimulatory Molecules CD80/86 Trigger Non-Specific Cytotoxic Cell of Nile tilapia (Oreochromis niloticus) to Kill CIK Cells" Fishes 7, no. 6: 353. https://doi.org/10.3390/fishes7060353
APA StyleHuang, Y., Chen, Z., Xie, R., Amoah, K., Wang, B., Cai, J., Lu, Y., & Jian, J. (2022). Costimulatory Molecules CD80/86 Trigger Non-Specific Cytotoxic Cell of Nile tilapia (Oreochromis niloticus) to Kill CIK Cells. Fishes, 7(6), 353. https://doi.org/10.3390/fishes7060353







