New Record of Saurida micropectoralis Shindo & Yamada, 1972 (Aulopiformes: Synodontidae) in the Southern Red Sea and Evidence of Range Expansion to East Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Analysis
2.2. Haplotype-Based ML Analysis
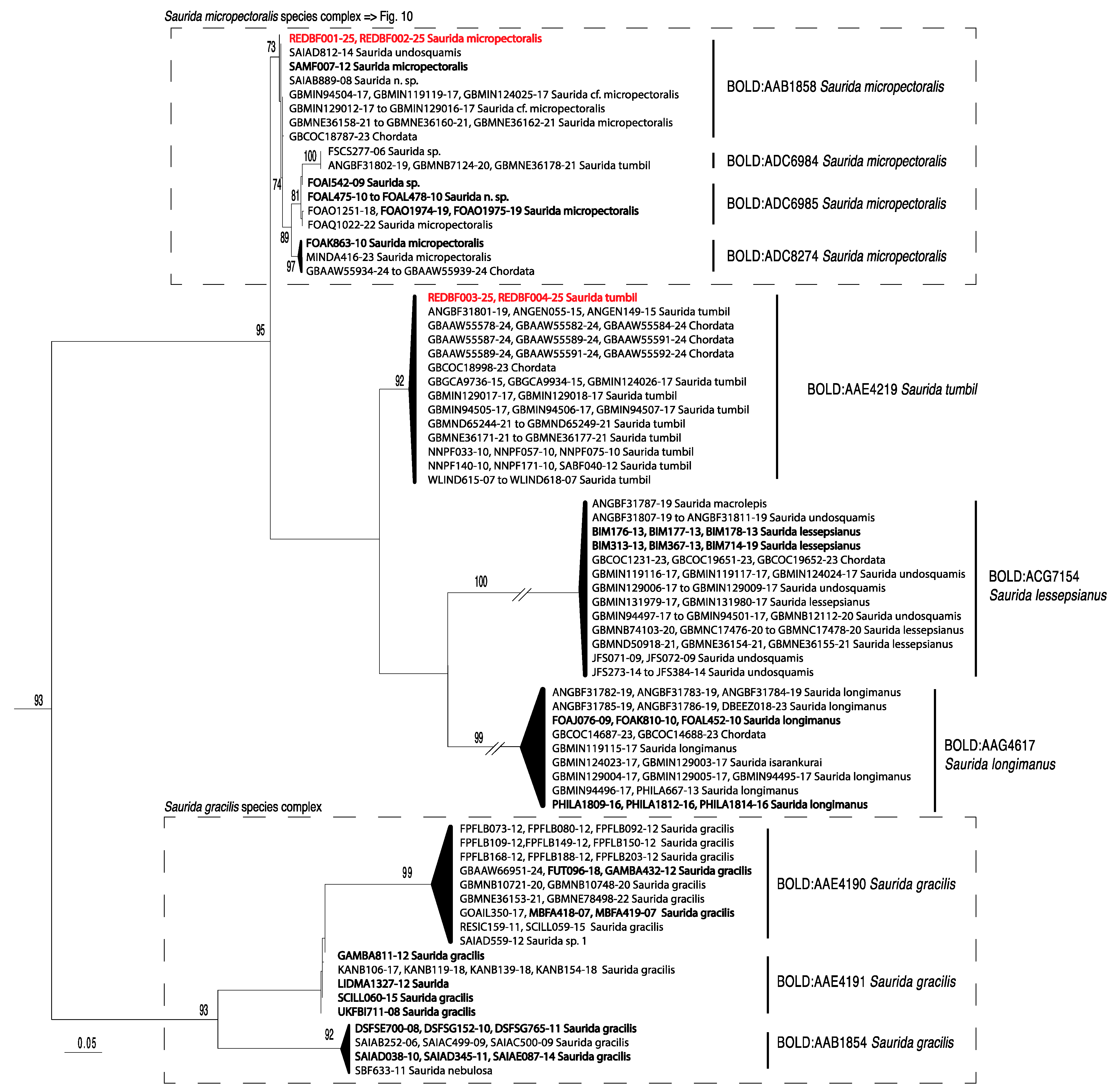
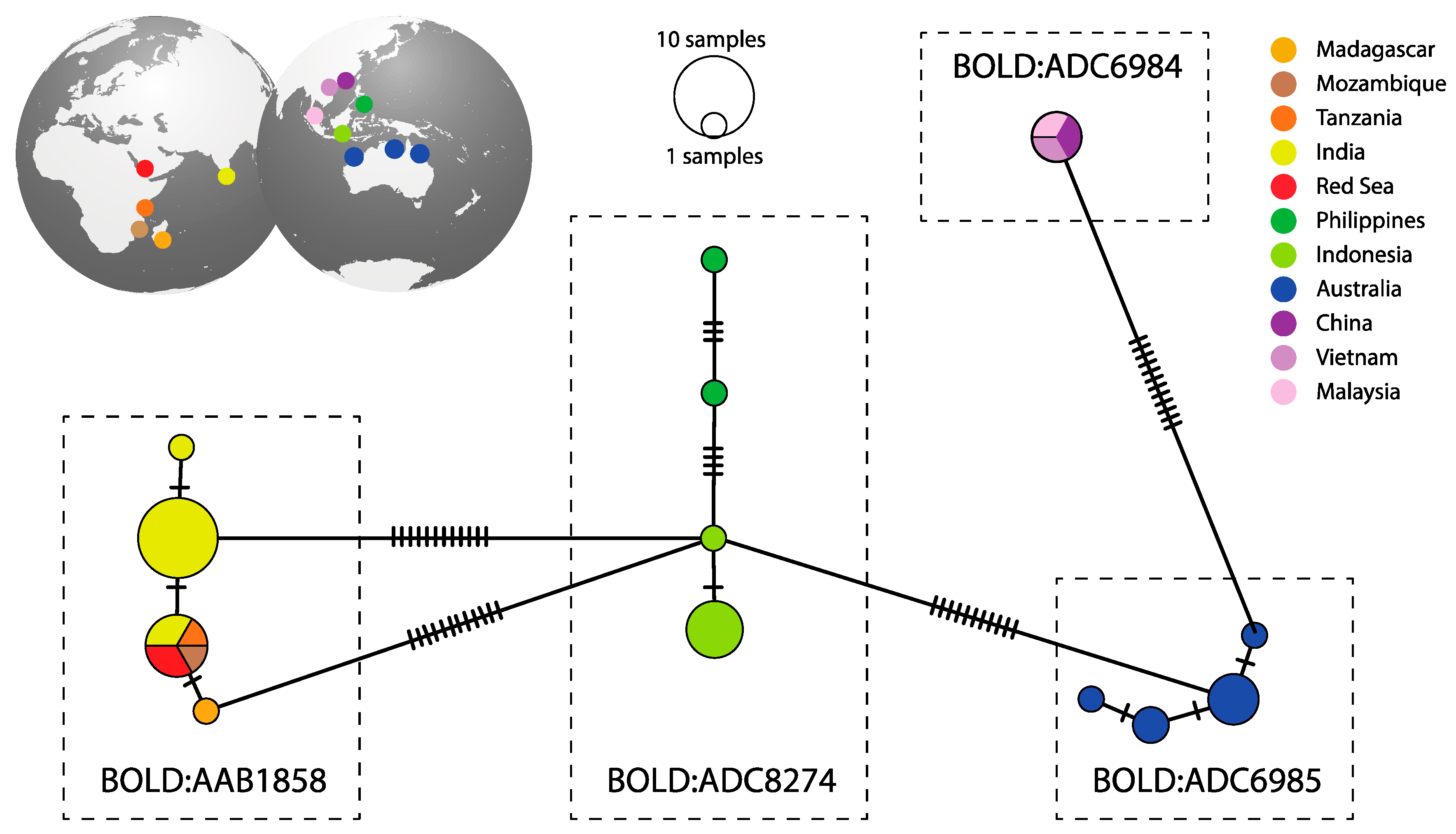
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fischer, W.; Whitehead, P.J.P. (Eds.) FAO Species Identification Sheets for Fishery Purposes. In Eastern Indian Ocean (Fishing Area 57) and Western Central Pacific (Fishing Area 71); FAO: Rome, Italy, 1974; Volumes 1–4. [Google Scholar]
- Fischer, W.; Bianchi, G. (Eds.) FAO Species Identification Sheets for Fishery Purposes. In Western Indian Ocean (Fishing Area 51); FAO: Rome, Italy, 1984; Volumes 1–6. [Google Scholar]
- Russell, B.C. Saurida golanii, a new deep-water lizardfish (Pisces: Synodontidae) from the Gulf of Aqaba, northern Red Sea. Zootaxa 2011, 3098, 21–25. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.N.; van der Laan, R. (Eds.) Eschmeyer’s Catalog of Fishes: Genera, Species, References. Electronic version. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 28 July 2025).
- Russell, B.C.; Malay, M.C.D.; Cabebe-Barnuevo, R.A. A New Deep-water Species of Saurida (Pisces: Synodontidae) from the South China Sea and Central Philippines. Zool. Stud. 2024, 63, 40. [Google Scholar]
- Bogorodsky, S.V.; Alpermann, T.J.; Mal, A.O.; Gabr, M.H. Survey of demersal fishes from southern Saudi Arabia, with five new records for the Red Sea. Zootaxa 2014, 3852, 401–437. [Google Scholar] [CrossRef]
- Russell, B.C.; Golani, D.; Tikochinski, Y. Saurida lessepsianus a new species of lizardfish (Pisces: Synodontidae) from the Red Sea and Mediterranean Sea, with a key to Saurida species in the Red Sea. Zootaxa 2015, 3956, 559–568. [Google Scholar] [CrossRef]
- Golani, D.; Fricke, R. Checklist of the Red Sea Fishes with delineation of the Gulf of Suez, Gulf of Aqaba, endemism and Lessepsian migrants. Zootaxa 2018, 4509, 215. [Google Scholar] [CrossRef]
- Williams, C.T.; Arostegui, M.C.; Braun, C.D.; Gaube, P.; Shriem, M.; Berumen, M.L. First records of two large pelagic fishes in the Red Sea: Wahoo (Acanthocybium solandri) and striped marlin (Kajikia audax). J. Mar. Biol. Assoc. U.K. 2022, 102, 505–508. [Google Scholar] [CrossRef]
- Goutham-Bharathi, M.P.; Sirajudheen, T.K.; Santucci, R.G.; Fricke, R.; Dimech, M. First record of Triacanthidae Bleeker, 1859 (Actinopterygii: Tetraodontiformes) from the Red Sea. Acta Ichthyol. Piscat. 2024, 54, 21–25. [Google Scholar] [CrossRef]
- Mohanraj, T.; Rajathy, T.J.; Kumar, T.A. New distributional record of the endemic Red Sea Parrotfish Scarus collana (Rüppell, 1835) from the southern coast of India. Zootaxa 2025, 5590, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Shindo, S.; Yamada, U. Description of three new species of the lizardfish genus Saurida, with a key to its Indo-Pacific species. UO (Jpn. Soc. Ichthyol.) 1972, 12, 1–4. [Google Scholar]
- Kadengal, S.T.; Ceyhan, T.; Tosunoğlu, Z.; Gireesh, S.; Charles, S.K.; Santucci, R.G.; Adam, A.M.S.; Tıraşın, E.M.; Ünal, V.; Dimech, M. Toward Sustainable Fisheries: Assessing Catch per Unit Effort, Retained Bycatch, and Discard Ratios in the Red Sea Shrimp Trawl Fishery of the Kingdom of Saudi Arabia. Sustainability 2024, 16, 10285. [Google Scholar] [CrossRef]
- Inoue, T.; Nakabo, T. The Saurida undosquamis group (Aulopiformes: Synodontidae), with description of a new species from southern Japan. Ichthyol. Res. 2006, 53, 379–397. [Google Scholar] [CrossRef]
- Shorthouse, D.P. SimpleMappr, an Online Tool to Produce Publication-Quality Point Maps. Available online: https://www.simplemappr.net (accessed on 28 July 2025).
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D.N. DNA Barcoding Australia’s fish species. Philos. Trans. R. Soc. 2005, B360, 1847–1857. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- GBIF Occurrence Download. Available online: https://doi.org/10.15468/dl.7ra9ax (accessed on 28 July 2025).
- Bogorodsky, S.V.; Randall, J.E. Endemic fishes of the Red Sea. In Oceanographic and Biological Aspects of the Red Sea; Rasul, N.M.A., Stewart, I.C.F., Eds.; Springer Oceanography: Cham, Switzerland, 2019; pp. 239–265. [Google Scholar]
- Silpa, S.; Srihari, M.; Pavan-Kumar, A.; Roul, S.K.; Russell, B.C.; Jaiswar, A.K. Mistaken by dots: Revealing the misidentification of Saurida lessepsianus (Actinopterygii: Aulopiformes: Synodontidae) along the west coast of India (eastern Arabian Sea). Acta Ichthyol. Piscat. 2021, 51, 185–191. [Google Scholar] [CrossRef]
- Zohra, K.; Imran, H.; Osmany, H. Record of Saurida lessepsianus, Russell, Golani and Tikochinski, 2015, (Order Aulopiformes; Family Synodontidae) from Pakistan (Northern Arabian Sea). Int. J. Biol. Biotech. 2022, 19, 355–361. [Google Scholar]
- Russell, B.C.; Houston, W. Offshore fishes of the Arafura Sea. Beagle Rec. North. Territ. Mus. Arts Sci. 1989, 6, 69–84. [Google Scholar] [CrossRef]
- Dutt, S.; Sagar, J.V. Saurida pseudotumbil-A new species of lizardfish (Teleostei: Synodidae) from Indian coastal waters. Indian Natl. Sci. Acad. 1981, B47, 845–851. [Google Scholar]
- Miyahara, H.; Choi, Y.; Yabe, M.; Nakaya, K. First record of a synodontid fish, Saurida micropectoralis from Japan. Jpn. J. Ichthyol. 2002, 49, 127–131. (In Japanese) [Google Scholar]
- Russell, B.C. Family Synodontidae. In Coastal Fishes of the Western Indian Ocean Volume 2; Heemstra, P.C., Heemstra, E., Ebert, D.A., Holleman, W., Randall, J.E., Eds.; South African Institute for Aquatic Biodiversity: Makhanda, South Africa, 2022; 614p. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]



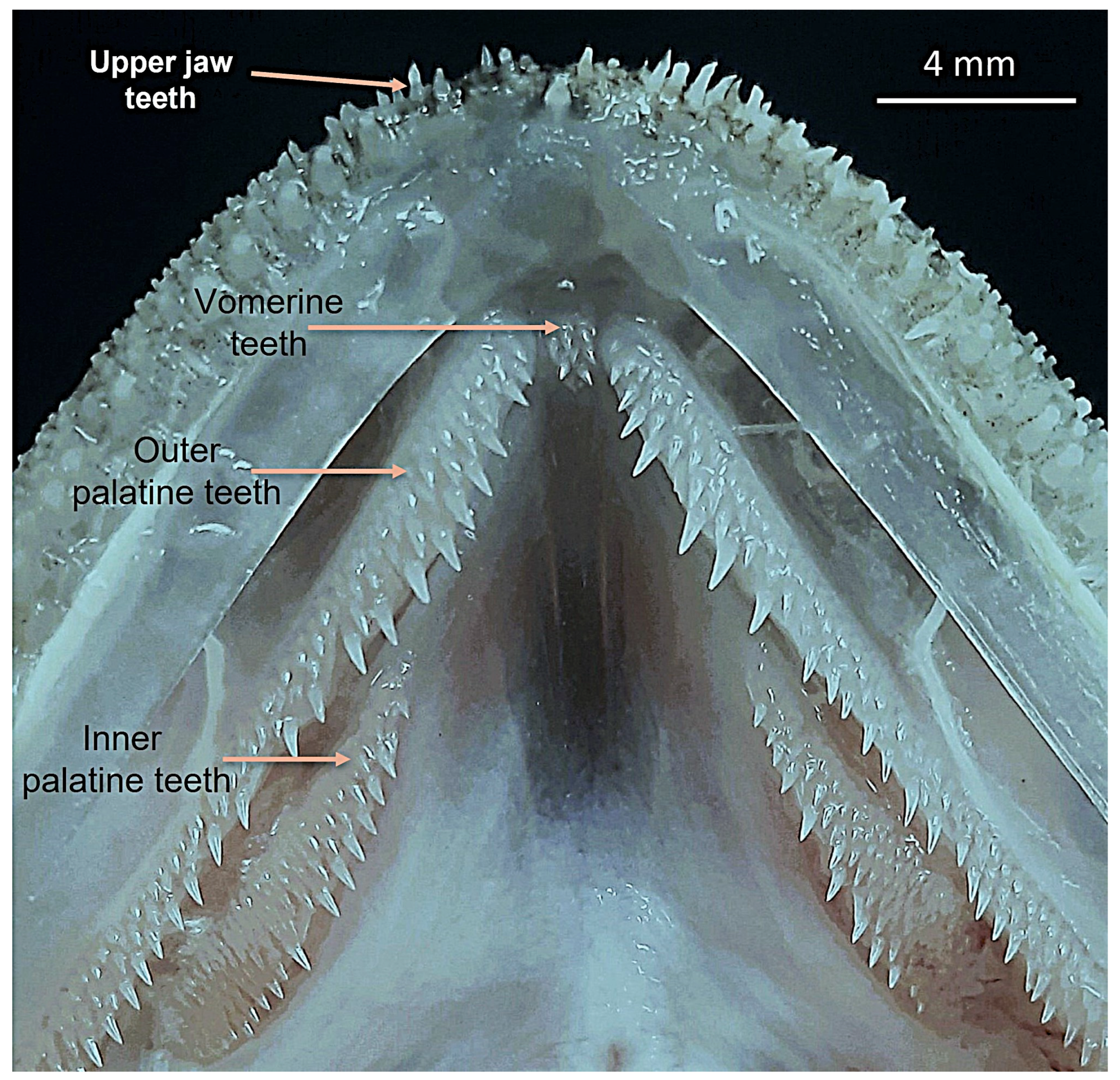

| Morphometrics (% Standard Length) | |||
|---|---|---|---|
| Min | Max | Mean ± SD | |
| Predorsal length | 40.0 | 43.3 | 41.8 ± 0.8 |
| Preadipose length | 79.3 | 83.1 | 81.2 ± 0.9 |
| Preanal length | 69.3 | 72.2 | 70.9 ± 0.9 |
| Preanal fin length | 73.2 | 76.3 | 74.7 ± 0.7 |
| Prepectoral length | 21.1 | 24.1 | 22.5 ± 0.8 |
| Prepelvic length | 34.6 | 37.7 | 36.2 ± 0.8 |
| Head length | 22.0 | 23.7 | 22.6 ± 0.7 |
| Body depth | 12.8 | 15.0 | 14.2 ± 0.5 |
| Body width | 12.9 | 16.6 | 14.6 ± 1.1 |
| Interpelvic width | 9.4 | 10.3 | 9.7 ± 0.4 |
| Pectoral fin length | 10.7 | 13.3 | 11.9 ± 0.7 |
| Pelvic fin length | 14.6 | 17.3 | 16.5 ± 0.8 |
| Length of 2nd dorsal ray | 16.3 | 20.9 | 18.1 ± 1.3 |
| Length of last dorsal ray | 3.2 | 7.5 | 4.6 ± 0.8 |
| Length of dorsal fin base | 11.9 | 13.8 | 12.6 ± 0.5 |
| Length of 2nd anal ray | 8.6 | 11.0 | 10.0 ± 0.6 |
| Length of last anal ray | 4.0 | 5.9 | 5.1 ± 0.6 |
| Length of anal fin base | 7.8 | 9.8 | 8.8 ± 0.6 |
| Length of caudal peduncle | 14.6 | 16.9 | 15.9 ± 0.9 |
| Depth of caudal peduncle | 5.4 | 6.4 | 5.9 ± 0.3 |
| Width of caudal peduncle | 5.7 | 7.0 | 6.5 ± 0.3 |
| Morphometrics: % head length and (%SL) | |||
| Snout length | 17.9 (3.6) | 29.8 (5.7) | 22.2 ± 3.3 (4.7 ± 0.6) |
| Eye diameter | 15.8 (3.5) | 25.5 (4.5) | 18.9 ± 2.5 (4.0 ± 0.3) |
| Snout width | 25.9 (6.4) | 35.7 (8.1) | 32.3 ± 2.6 (7.1 ± 0.4) |
| Interorbital width | 17.6 (4.6) | 25.2 (5.6) | 22.9 ± 1.8 (5.1 ± 0.3) |
| Postorbital length | 57.4 (12.5) | 64.3 (14.6) | 62.3 ± 2.0 (13.7 ± 0.6) |
| Upper jaw length | 56.9 (12.7) | 75.2 (16.5) | 67.5 ± 4.5 (15.0 ± 1.2) |
| Meristics | Min | Max | Mode |
| No. of dorsal fin rays | 11 | 12 | 12 |
| No. of pectoral fin rays | 14 | 15 | 14 |
| No. of pelvic fin rays | 9 | 9 | 9 |
| No. of anal fin rays | 10 | 12 | 11 |
| Pored lateral-line scales | 56 | 59 | 57 |
| Transverse scales above lateral line | 4.5 | 4.5 | 4.5 |
| Transverse scales below lateral line | 5.5 | 5.5 | 5.5 |
| Scale rows below LL with melanophores | 2 | 2 | 2 |
| Predorsal scales | 19 | 22 | 20 |
| Preadipose scales | 16 | 19 | 18 |
| Postadipose scales | 10 | 12 | 11 |
| No. rows of palatine teeth anteriorly | 3 | 4 | 3 |
| No. vomerine teeth | 2 | 10 | 6 |
| Present Study (26 Specimens: 202–315 mm SL) | Miyahara et al., 2002 [30] (1 Specimen: 324 mm SL) | Shindo and Yamada, 1972 [12] (32 Specimens: 86–246 mm SL) | |
|---|---|---|---|
| Morphometrics: As a percentage of standard length | |||
| Predorsal length | 40.0–43.3 | 43.4 | 42.0–44.9 |
| Preanal fin length | 73.2–76.3 | 75.9 | 75.6–79.7 |
| Prepelvic length | 34.6–37.7 | 37.6 | 36.7–40.5 |
| Head length | 22.0–23.7 | 23.7 | 24.1–26.7 |
| Body depth | 12.8–15.0 | 14.8 | 11.5–15.8 |
| Pectoral fin length | 10.7–13.3 | 12.0 | 10.8–12.9 |
| Pelvic fin length | 14.6–17.3 | 16.5 | 14.8–17.4 |
| Length of longest dorsal ray | 16.3–20.9 | 20.1 | 18.8–22.9 |
| Length of dorsal fin base | 11.9–13.8 | 14.0 | - |
| Length of longest anal ray | 8.6–11.0 | 10.4 | - |
| Length of anal fin base | 7.8–9.8 | 8.7 | - |
| Length of caudal peduncle | 14.6–16.9 | 15.3 | - |
| Depth of caudal peduncle | 5.4–6.4 | 6.3 | - |
| Snout length | 3.6–5.7 | 5.8 | - |
| Eye diameter | 3.5–4.5 | 3.8 | 3.66–5.46 |
| Interorbital width | 4.6–5.6 | 5.4 | - |
| Upper jaw length | 12.7–16.5 | 16.2 | - |
| Meristics | |||
| No. of dorsal fin rays | 11–12 | 12 | 11–12 |
| No. of pectoral fin rays | 14–15 | 15 | 14–15 |
| No. of pelvic fin rays | 9 | 9 | 9 |
| No. of anal fin rays | 10–12 | 11 | 10–11 |
| Pored lateral-line scales | 56–59 | 55 | 56–58 |
| Transverse scales above LL | 4.5 | 5 | 5 |
| Transverse scales below LL | 5.5 | 6 | - |
| Predorsal scales | 20–22 | 22 | 20–21 |
| Character | S. golanii | S. lessepsianus | S. longimanus | S. micropectoralis | S. tumbil |
|---|---|---|---|---|---|
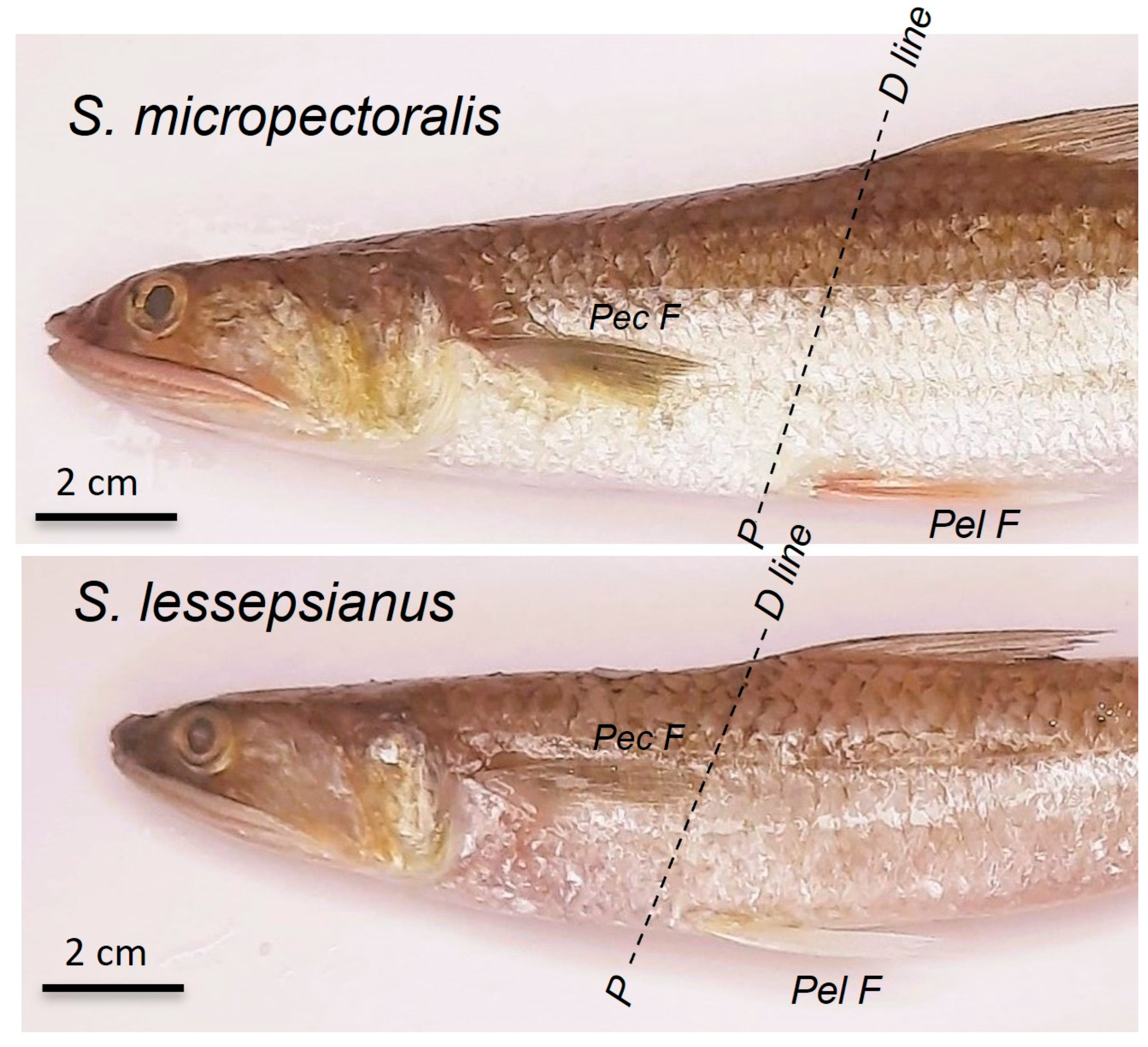
| Position of the pectoral fin versus pelvic fin origin | Largely beyond | Slightly beyond | Well beyond | Not reaching | Just reaching |
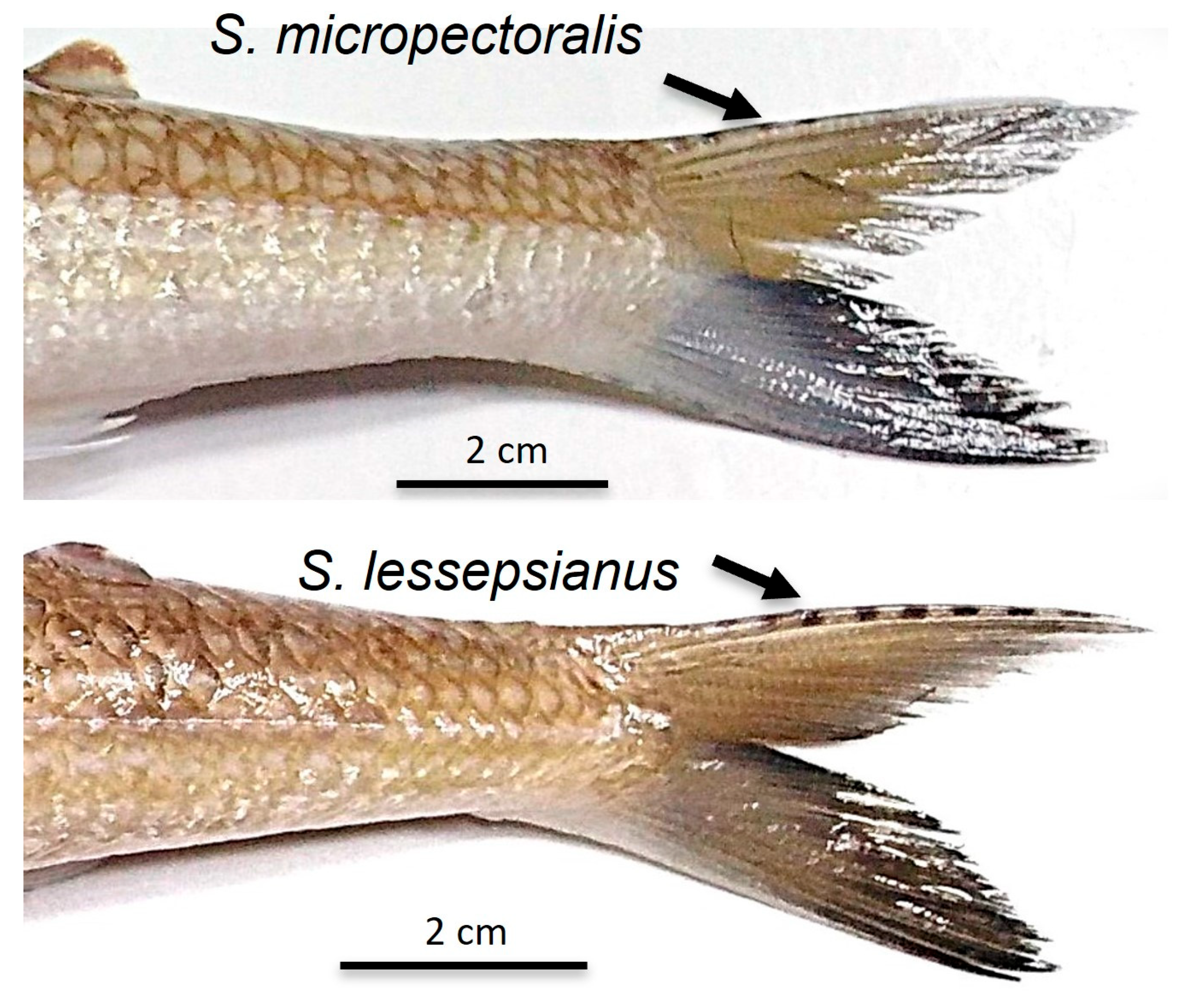
| Bars on upper edges of caudal fin | Absent | Present (black spots) | Present (indistinct) | Present (faint dusky spots) | Absent |
| Blotches on flanks | Absent | Present | Present | Present | Absent |
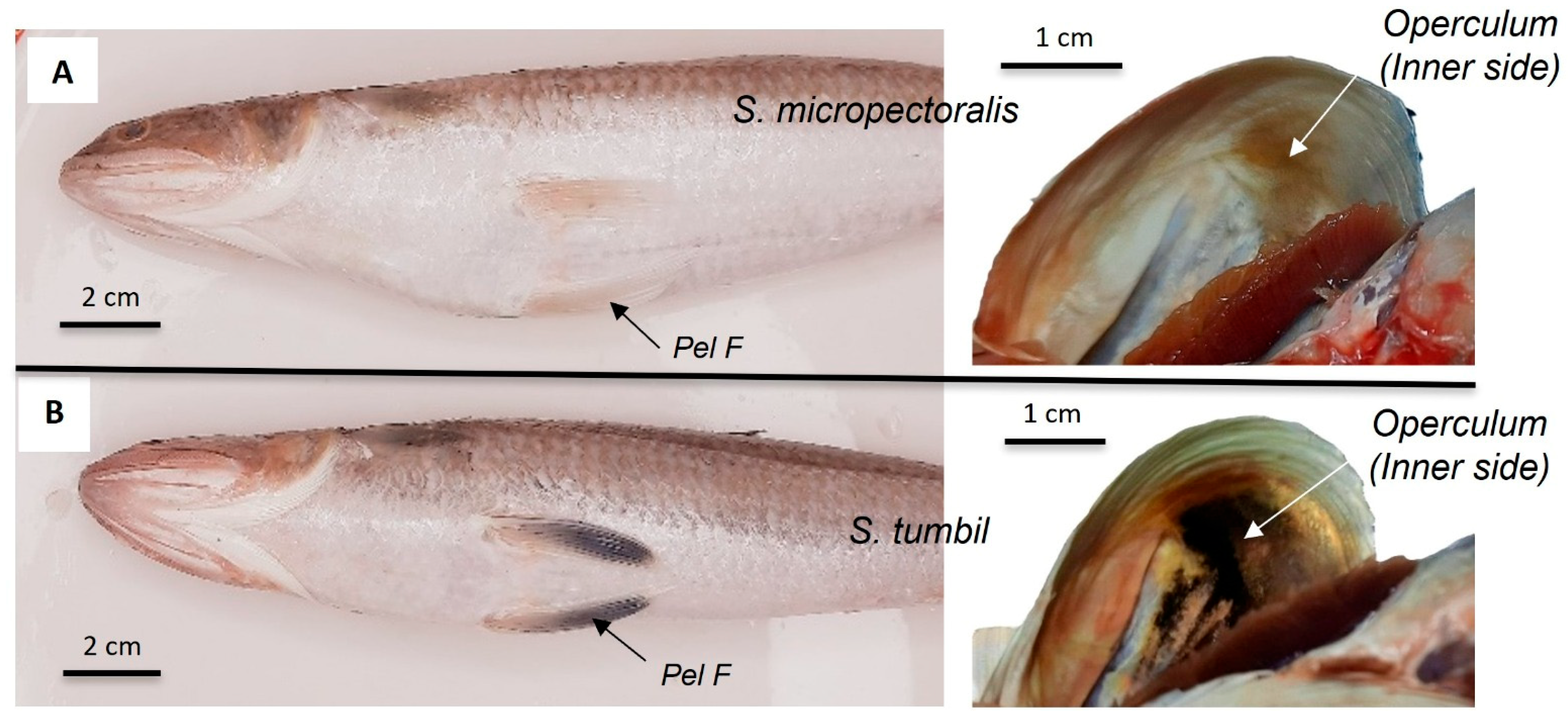
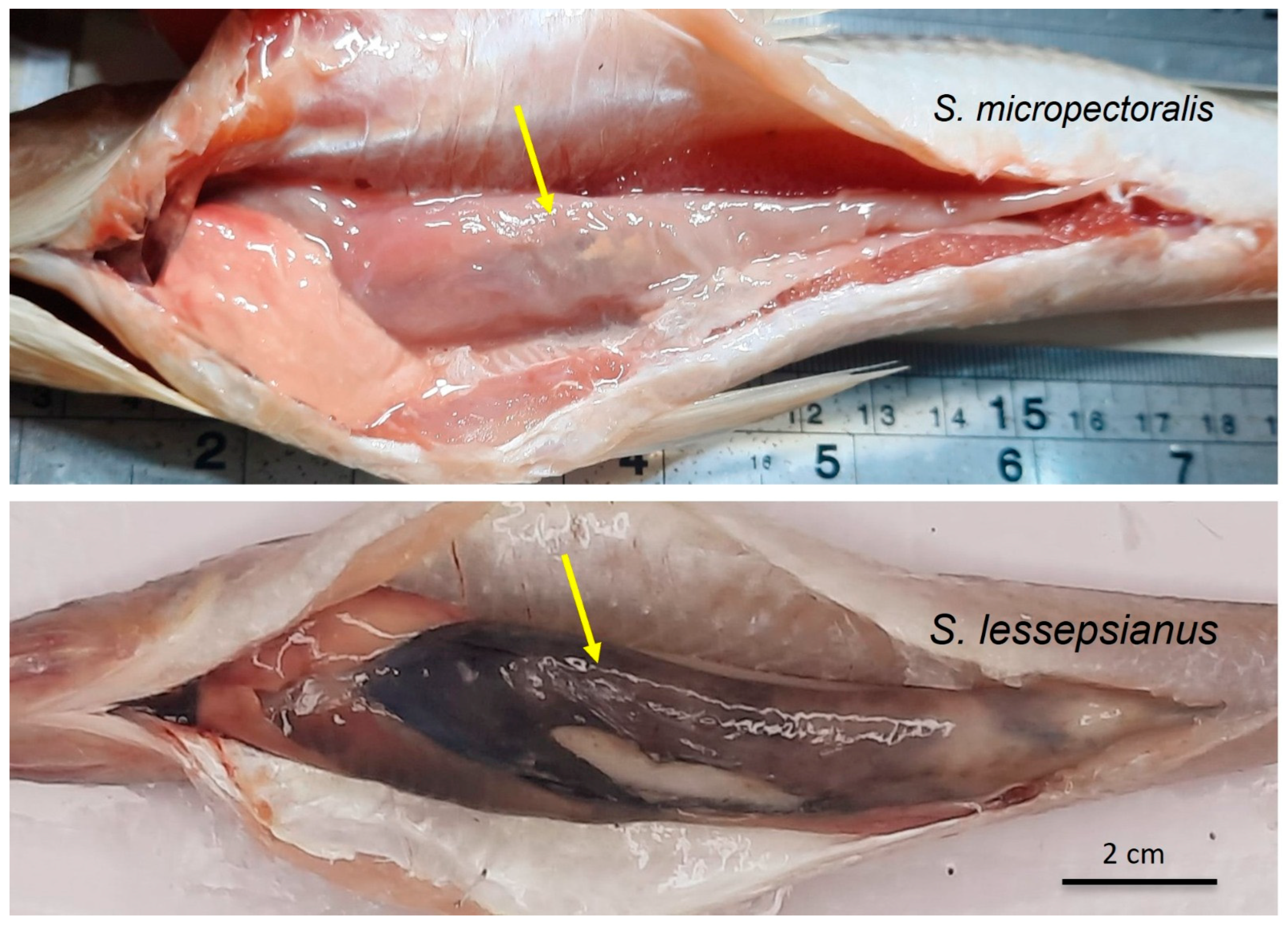
| Stomach color | Dark anteriorly | Grey to dark anteriorly | Dark | Unpigmented | Unpigmented |
| Intestine color | Black | Unpigmented | Striped | Unpigmented | Unpigmented |
| Pelvic fin color | Unpigmented | Unpigmented | Unpigmented | Unpigmented | Dark |
| Outer series of palatine teeth | Three rows | Two rows | Two rows | Three rows | Three rows |
| No. of lateral-line scales | 53–56 | 47–51 | 46–50 | 56–59 | 55–58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabr, M.H.; Abu El-Regal, M.A.; El-Sherbiny, M.M.; Al-Harby, M.A.; Durand, J.-D. New Record of Saurida micropectoralis Shindo & Yamada, 1972 (Aulopiformes: Synodontidae) in the Southern Red Sea and Evidence of Range Expansion to East Africa. Fishes 2025, 10, 452. https://doi.org/10.3390/fishes10090452
Gabr MH, Abu El-Regal MA, El-Sherbiny MM, Al-Harby MA, Durand J-D. New Record of Saurida micropectoralis Shindo & Yamada, 1972 (Aulopiformes: Synodontidae) in the Southern Red Sea and Evidence of Range Expansion to East Africa. Fishes. 2025; 10(9):452. https://doi.org/10.3390/fishes10090452
Chicago/Turabian StyleGabr, Mohamed Hosny, Mohamed Ahmed Abu El-Regal, Mohsen Mohamed El-Sherbiny, Mamdouh Aly Al-Harby, and Jean-Dominique Durand. 2025. "New Record of Saurida micropectoralis Shindo & Yamada, 1972 (Aulopiformes: Synodontidae) in the Southern Red Sea and Evidence of Range Expansion to East Africa" Fishes 10, no. 9: 452. https://doi.org/10.3390/fishes10090452
APA StyleGabr, M. H., Abu El-Regal, M. A., El-Sherbiny, M. M., Al-Harby, M. A., & Durand, J.-D. (2025). New Record of Saurida micropectoralis Shindo & Yamada, 1972 (Aulopiformes: Synodontidae) in the Southern Red Sea and Evidence of Range Expansion to East Africa. Fishes, 10(9), 452. https://doi.org/10.3390/fishes10090452








