DNA Barcode and Correct Scientific Name of Golden Pompano, an Important Marine Aquaculture Fish Species in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Molecular Identification
| Species | Locality | Bold Bin | GenBank Accession No. | Citation |
| Trachinotus africanus Smith, 1967 | South Africa | TZMSC062-05 | JF494716 | Steinke et al., 2016 [24] |
| Indonesia, Java | FOAH845-08 | N/A | N/A | |
| Trachinotus anak Ogilby, 1909 | Australia | FOAC410-05 | EF609479 | Ward and Holmes, 2007 [25] |
| Australia | FOAC409-05 | N/A | N/A | |
| Trachinotus baillonii (Lacepède, 1801) | Australia | FOAC407-05 | EF609480 | Ward and Holmes, 2007 [25] |
| China, Zhoushan | GBGCA9146-15 | KM245967 | Xu et al., 2015 [26] | |
| Trachinotus blochii (Lacepède, 1801) | Australia | FOAC411-05 | EF609481 | Ward and Holmes, 2007 [25] |
| Seychelles | UKFBJ917-08 | KF930508 | N/A | |
| China | GBMNA14603-19 | KJ184305 | Zhang et al., 2016 [27] | |
| Australia | LIFS650-08 | KP194696 | Steinke et al., 2017 [28] | |
| Trachinotus botla (Shaw, 1803) | Indonesia | FOAJ894-09 | GU673700 | N/A |
| South Africa | TZMSC524-05 | JF494718 | Steinke et al., 2016 [24] | |
| South Africa | TZMSA220-04 | JF494722 | Steinke et al., 2016 [24] | |
| Trachinotus carolinus (Linnaeus, 1766) | Brazil, Sao Paulo | MFSP926-11 | JQ365599 | Ribeiro et al., 2012 [29] |
| N/A | GBMNA14604-19 | KJ556976 | Zhang et al., 2016 [30] | |
| United States, Texas | GBMNE87372-22 | ON995519 | N/A | |
| Trachinotus coppingeri Günther, 1884 | Australia | FOAC406-05 | EF609482 | Ward and Holmes, 2007 [25] |
| Australia | FOAD511-05 | N/A | N/A | |
| Trachinotus falcatus (Linnaeus, 1758) | Brazil, Sao Paulo | MFSP394-10 | JQ365601 | Ribeiro et al., 2012 [29] |
| Mexico | N/A | KR086928 | N/A | |
| Mexico | MLIII196-08 | N/A | Leyva-Cruz et al., 2016 [31] | |
| Trachinotus goodei Jordan and Evermann, 1896 | Brazil, Sao Paulo | MFSP145-09 | GU702385 | Ribeiro et al., 2012 [29] |
| Trachinotus goreensis Cuvier, 1832 | Nigeria | BAFEN355-10 | HM883015 | Nwani et al., 2011 [32] |
| Trachinotus maxillosus Cuvier, 1832 | Gambia, Tanji | N/A | KX512710 | Damerau et al., 2018 [33] |
| Trachinotus mookalee Cuvier, 1832 | India | GBMIN119993-17 | KU296860 | N/A |
| India | GBMIN130195-17 | KU296861 | N/A | |
| India | GBMIN124842-17 | KU296862 | N/A | |
| Trachinotus ovatus (Linnaeus, 1758) | Turkey | DNATR106-12 | JQ624009 | N/A |
| Turkey | DNATR1776-13 | KC501749 | Keskin and Atar, 2013 [13] | |
| Portugal | MLFPI019-09 | KJ768314 | Landi et al., 2014 [14] | |
| Israel | BIM405-15 | N/A | N/A | |
| Trachinotus ovatus (Linnaeus, 1758) | China, Zhoushan | GBGCA9147-15 | KM245969 | Xu et al., 2015 [26] |
| China, South China Sea | FSCS483-07 | EU595325 | Zhang and Hanner, 2012 [11] | |
| China, South China Sea | ANGBF5359-12 | JN242717 | Zhang and Hanner, 2012 [11] | |
| China | GBMNA14602-19 | KF356397 | Xie et al., 2015 [34] | |
| Central China Market | N/A | KP112476 | Shen et al., 2016 [35] | |
| N/A | ANGBF17718-19 | KY802069 | Muri et al., 2018 [36] | |
| Vietnam | ANGBF55875-19 | MK227447 | N/A | |
| China, Beibu Gulf | GBMNE78207-22 | OP247575 | N/A | |
| Trachinotus paitensis Cuvier, 1832 | United States, California | ANGBF1237-12 | HQ010062 | N/A |
| Peru | GBMND24296-21 | MN880583 | Marín et al., 2021 [37] | |
| Peru | GBMND24300-21 | MN880587 | Marín et al., 2021 [37] | |
| Trachinotus rhodopus Gill, 1863 | Costa Rica | RDFCA397-05 | N/A | N/A |
| Trachinotus stilbe (Jordan and McGregor, 1898) | Ecuador, Galapagos | LIDMA1275-12 | N/A | N/A |
| Lichia amia (Linnaeus, 1758) | Turkey | N/A | KY176514 | N/A |
| Turkey | N/A | JQ623944 | N/A |
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, D.C.; Guo, L.; Guo, H.Y.; Zhu, K.C.; Li, S.Q.; Zhang, Y.; Zhang, N.; Liu, B.S.; Jiang, S.G.; Li, J.T. Chromosome-level genome assembly of golden pompano (Trachinotus ovatus) in the family Carangidae. Sci. Data. 2019, 6, 216. [Google Scholar] [CrossRef]
- Liu, B.; Guo, H.Y.; Zhu, K.C.; Guo, L.; Liu, B.S.; Zhang, N.; Yang, J.W.; Jiang, S.G.; Zhang, D.C. Growth, physiological, and molecular responses of golden pompano Trachinotus ovatus (Linnaeus, 1758) reared at different salinities. Fish Physiol. Biochem. 2019, 45, 1879–1893. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Agriculture and Rural Affairs of People’s Republic of China, National Fisheries Technology Extension Center, China Society of Fisheries. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2024; p. 22. (In Chinese) [Google Scholar]
- San, L.Z.; Liu, B.S.; Liu, B.; Guo, H.Y.; Guo, L.; Zhang, N.; Zhu, K.C.; Jiang, S.G.; Zhang, D.C. Transcriptome analysis of gills provides insights into translation changes under hypoxic stress and reoxygenation in Golden Pompano, Trachinotus ovatus (Linnaeus 1758). Front. Mar. Sci. 2021, 8, 763622. [Google Scholar] [CrossRef]
- Liu, M.; Chen, X.; Yang, S.Y. Marine Fishes of Southern Fujian, China; China Ocean Press: Beijing, China, 2014; pp. 165–166. (In Chinese) [Google Scholar]
- Wu, H.L.; Zhong, J.S. Key to Marine and Estuarial Fishes of China; China Agriculture Press: Beijing, China, 2021; pp. 709–711. (In Chinese) [Google Scholar]
- Fan, B.; Xie, D.Z.; Li, Y.W.; Wang, X.L.; Qi, X.; Li, S.S.; Meng, Z.N.; Chen, X.H.; Peng, J.Y.; Yang, Y.J.; et al. A single intronic single nucleotide polymorphism in splicing site of steroidogenic enzyme hsd17b1 is associated with phenotypic sex in oyster pompano, Trachinotus anak. Proc. R. Soc. B 2021, 288, 20212245. [Google Scholar] [CrossRef] [PubMed]
- Linnaeus, C. Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis, 10th ed.; Laurentius Salvius: Stockholm, Sweden, 1758; pp. 295–297. (In Latin) [Google Scholar]
- Kendall, W.C.; Goldsborough, E.L. The shore fishes. Mem. Mus. Comp. Zool. Harvard Coll. 1911, 26, 269–271. [Google Scholar]
- Zhang, Q.; Hong, W.S.; Shao, K.T. Studies on taxonomic characters of Trachinotus ovatus and Trachinotus blochii from net cage mariculture. J. Oceanogr. Taiwan Strait 2000, 19, 499–506. (In Chinese) [Google Scholar] [CrossRef]
- Zhang, J.B.; Hanner, R. Molecular approach to the identification of fish in the South China Sea. PLoS ONE 2012, 7, e30621. [Google Scholar] [CrossRef]
- Boldsystems. Available online: https://v3.boldsystems.org/index.php/Public_RecordView?processid=FSCS483-07 (accessed on 10 February 2025).
- Keskin, E.; Atar, H.H. DNA barcoding commercially important fish species of Turkey. Mol. Ecol. Resour. 2013, 13, 788–797. [Google Scholar] [CrossRef]
- Landi, M.; Dimech, M.; Arculeo, M.; Biondo, G.; Martins, R.; Carneiro, M.; Carvalho, G.R.; Brutto, S.L.; Costa, F.O. DNA Barcoding for Species Assignment: The Case of Mediterranean Marine Fishes. PLoS ONE 2014, 9, e106135. [Google Scholar] [CrossRef]
- Boldsystems. Available online: https://v3.boldsystems.org/index.php/Public_RecordView?processid=MLFPI019-09 (accessed on 10 February 2025).
- Fishbase. Available online: https://fishbase.mnhn.fr/summary/Trachinotus-ovatus.html (accessed on 10 February 2025).
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Heber, P.D.N. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Haeseler, A.V.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Zhang, J.J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Steinke, D.; Connell, A.D.; Hebert, P.D.N. Linking adults and immatures of South African marine fishes. Genome 2016, 59, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.D.; Holmes, B.H. An analysis of nucleotide and amino acid variability in the barcode region of cytochrome c oxidase I (cox1) in fishes. Mol. Ecol. Notes 2007, 7, 899–907. [Google Scholar] [CrossRef]
- Xu, Z.T.; Wang, H.P.; Gu, H.M.; Chen, Y.J. DNA barcoding Carangids in Putuo Sea. J. Zhejiang Ocean Univ. Nat. Sci. 2015, 34, 105–111. (In Chinese) [Google Scholar] [CrossRef]
- Zhang, D.C.; Wang, L.; Guo, H.Y.; Ma, Z.H.; Jiang, S.G. The complete mitochondrial genome of snubnose pompano Trachinotus blochii (Teleostei, Carangidae). Mitochondrial DNA Part A 2016, 27, 431–432. [Google Scholar] [CrossRef]
- Steinke, D.; deWaard, J.R.; Gomon, M.F.; Johnson, J.W.; Larson, H.K.; Lucanus, O.; Moore, G.I.; Reader, S.; Ward, R.D. DNA barcoding the fishes of Lizard Island (Great Barrier Reef). Biodivers. Data J. 2017, 5, e12409. [Google Scholar] [CrossRef]
- Ribeiro, A.O.; Caires, R.A.; Mariguela, T.C.; Pereira, L.H.G.; Hanner, R.; Oliveira, C. DNA barcodes identify marine fishes of São Paulo State, Brazil. Mol. Ecol. Resour. 2012, 12, 1012–1020. [Google Scholar] [CrossRef]
- Zhang, D.C.; Wang, L.; Guo, H.Y.; Ma, Z.H.; Zhang, N.; Lin, J.D.; Jiang, S.G. Complete mitochondrial genome of Florida pompano Trachinotus carolinus (Teleostei, Carangidae). Mitochondrial DNA Part A 2016, 27, 597–598. [Google Scholar] [CrossRef]
- Leyva-Cruz, E.; Vásquez-Yeomans, L.; Carrillo, L.; Valdez-Moreno, M. Identifying pelagic fish eggs in the southeast Yucatan Peninsula using DNA barcodes. Genome 2016, 59, 1117–1129. [Google Scholar] [CrossRef]
- Nwani, C.D.; Becker, S.; Braid, H.E.; Ude, E.F.; Okogwu, O.I.; Hanner, R. DNA barcoding discriminates freshwater fishes from southeastern Nigeria and provides river system-level phylogeographic resolution within some species. Mitochondrial DNA 2011, 22, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Damerau, M.; Freese, M.; Hanel, R. Multi-gene phylogeny of jacks and pompanos (Carangidae), including placement of monotypic vadigo Campogramma glaycos. J. Fish Biol. 2018, 92, 190–202. [Google Scholar] [CrossRef]
- Xie, Z.Z.; Li, S.S.; Yao, M.; Lu, D.Q.; Li, Z.H.; Meng, Z.N.; Zhang, Y.; Lin, H.R. The complete mitochondrial genome of the Trachinotus ovatus (Teleostei, Carangidae). Mitochondrial DNA 2015, 26, 644–646. [Google Scholar] [CrossRef]
- Shen, Y.J.; Kang, J.L.; Chen, W.T.; He, S.P. DNA barcoding for the identification of common economic aquatic products in Central China and its application for the supervision of the market trade. Food. Control 2016, 61, 79–91. [Google Scholar] [CrossRef]
- Muri, C.D.; Vandamme, S.G.; Peace, C.; Barnes, W.; Mariani, S. Biodiversity defrosted: Unveiling non-compliant fish trade in ethnic food stores. Biol. Conserv. 2018, 217, 419–427. [Google Scholar] [CrossRef]
- Marín, A.; Wuest, R.G.; Grillo-Nuñez, J.; Alvarez-Jaque, I.B.; Riveros, J.C. DNA barcoding reveals overlooked shark and bony fish species in landing reports of small-scale fisheries from northern Peru. Mar. Fish. Sci. 2021, 35, 307–314. [Google Scholar] [CrossRef]
- Eschmeyer’s Catalog of Fishes. Available online: https://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatget.asp?spid=15462 (accessed on 10 February 2025).
- Hebert, P.D.N.; Ratnasingham, S.; de Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit I divergences among closely related species. Proc. R. Soc. B 2003, 270, S96–S99. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.T.; Chang, J.S.; Jeng, M.S.; Tu, T.H.; Chiu, Y.W.; Ho, C.W.; Chan, T.Y.; Ho, P.H.; Joung, S.J.; Chao, S.M.; et al. A Guide Book of Common Economic Aquatic Animals and Plants in Taiwan; Fisheries Agency, Council of Agriculture: Taipei, Taiwan, 2015; p. 311. (In Chinese) [Google Scholar]
- Du, T.; Luo, J. Study on techniques of artificial breeding of Trachinotus blochii. Mar. Fish. Res. 2004, 25, 46–50. (In Chinese) [Google Scholar] [CrossRef]
- Collins, R.A.; Cruickshank, R.H. The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 2013, 13, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Antil, S.; Abraham, J.S.; Sripoorna, S.; Maurya, S.; Dagar, J.; Makhija, S.; Bhagat, P.; Gupta, R.; Sood, U.; Lal, R.; et al. DNA barcoding, an effective tool for species identification: A review. Mol. Biol. Rep. 2023, 50, 761–775. [Google Scholar] [CrossRef]
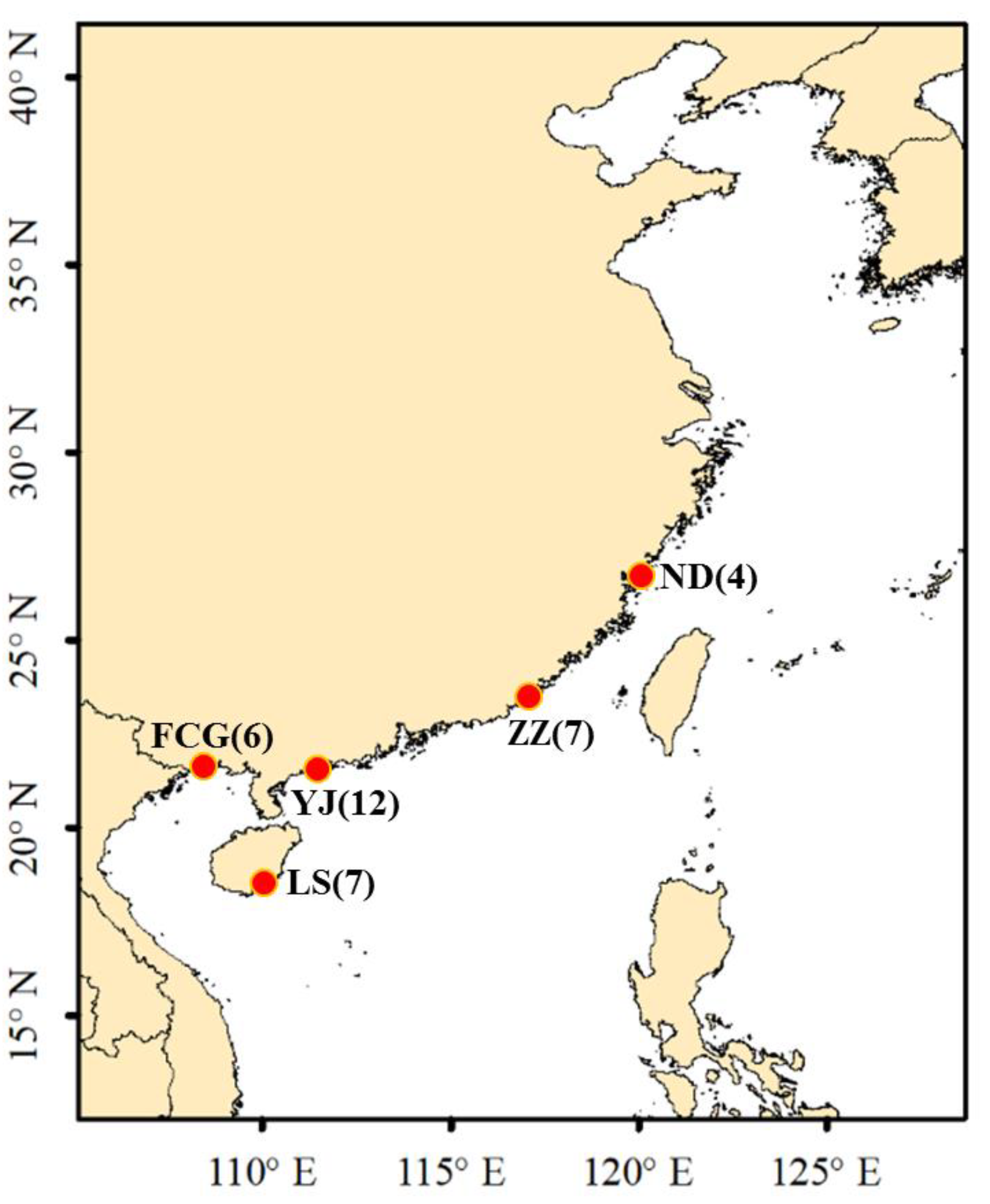
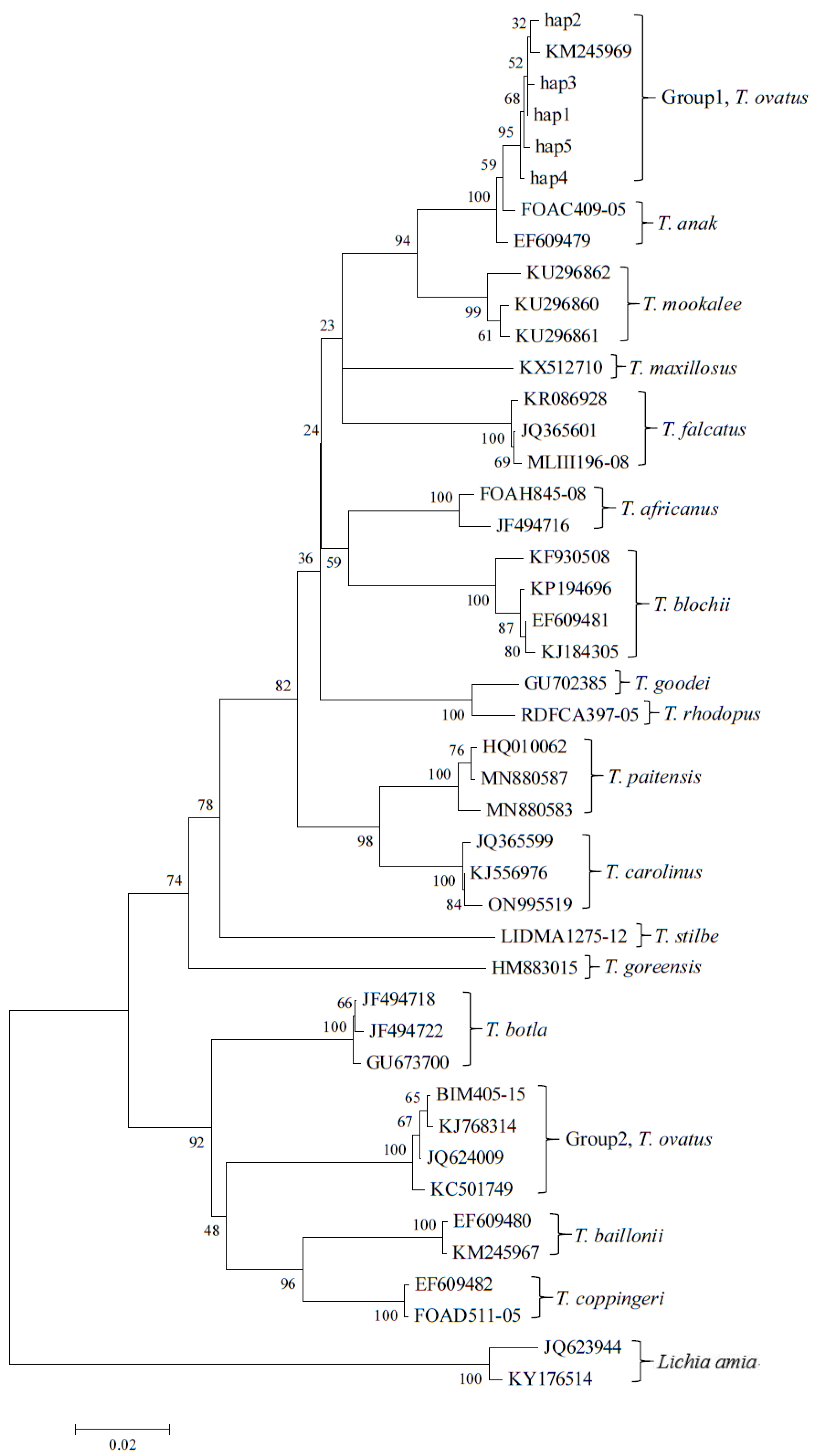

| Haplotype | Individual No. | GenBank Accession No. |
|---|---|---|
| hap1 | ZZ2, ZZ4, ZZ5, ZZ6, YJ1, YJ2, YJ3, YJ4, YJ5, YJ6, YJ7, YJ8, YJ9, YJ10, YJ12, LS1, LS3, LS4, LS5, LS6, LS7, ND2, ND4, FCG1, FCG2, FCG3, FCG5, EU595325, JN242717, KF356397, KP112476, OP247575 | PV164038 |
| hap2 | ZZ1, MK227447 | PV164039 |
| hap3 | YJ11 | PV164040 |
| hap4 | ZZ3, LS2, FCG4 | PV164041 |
| hap5 | FCG6, ZZ7, ND1, ND3, KY802069 | PV164042 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, A.; An, C.; Wang, H.; Che, S.; Liu, S.; Zhuang, Z. DNA Barcode and Correct Scientific Name of Golden Pompano, an Important Marine Aquaculture Fish Species in China. Fishes 2025, 10, 129. https://doi.org/10.3390/fishes10030129
Li A, An C, Wang H, Che S, Liu S, Zhuang Z. DNA Barcode and Correct Scientific Name of Golden Pompano, an Important Marine Aquaculture Fish Species in China. Fishes. 2025; 10(3):129. https://doi.org/10.3390/fishes10030129
Chicago/Turabian StyleLi, Ang, Changting An, Huan Wang, Shuai Che, Shufang Liu, and Zhimeng Zhuang. 2025. "DNA Barcode and Correct Scientific Name of Golden Pompano, an Important Marine Aquaculture Fish Species in China" Fishes 10, no. 3: 129. https://doi.org/10.3390/fishes10030129
APA StyleLi, A., An, C., Wang, H., Che, S., Liu, S., & Zhuang, Z. (2025). DNA Barcode and Correct Scientific Name of Golden Pompano, an Important Marine Aquaculture Fish Species in China. Fishes, 10(3), 129. https://doi.org/10.3390/fishes10030129






