Optimizing Cell Density and Unveiling Cytotoxic Profiles of DMSO and Ethanol in Six Cancer Cell Lines: Experimental and In Silico Insights
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Reagents
2.2. Cell Culture
2.3. Cell Seeding
2.4. MTT Assay Procedure
2.5. Calibration Curve and Data Analysis
2.6. Assessment of Solvent Cytotoxicity
2.7. Molecular Docking Studies
2.8. Intra-Molecular Interaction Studies
2.9. Statistical Analysis
3. Results
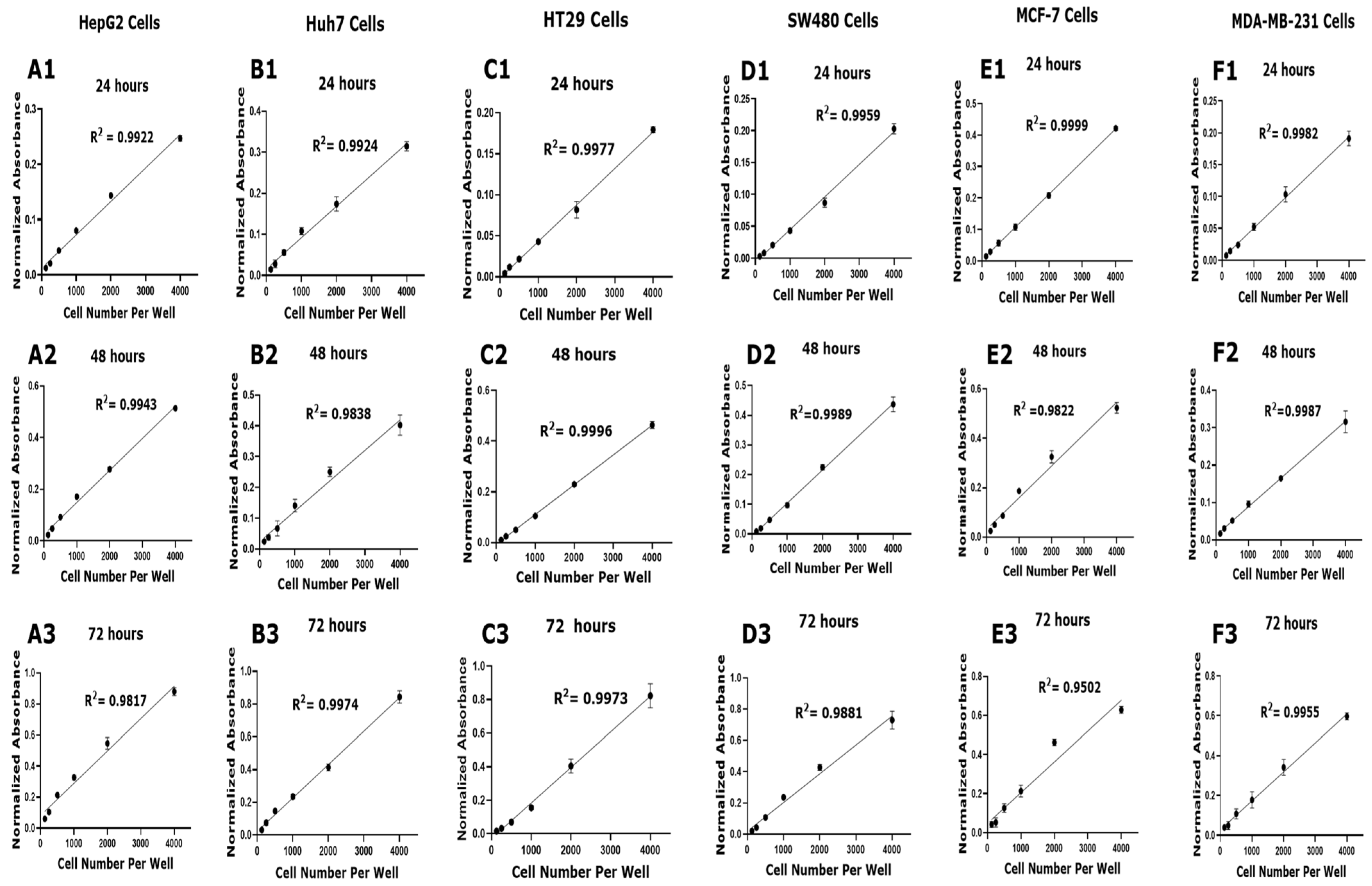
3.1. Optimization of Cell Density for Accurate Viability Assessment
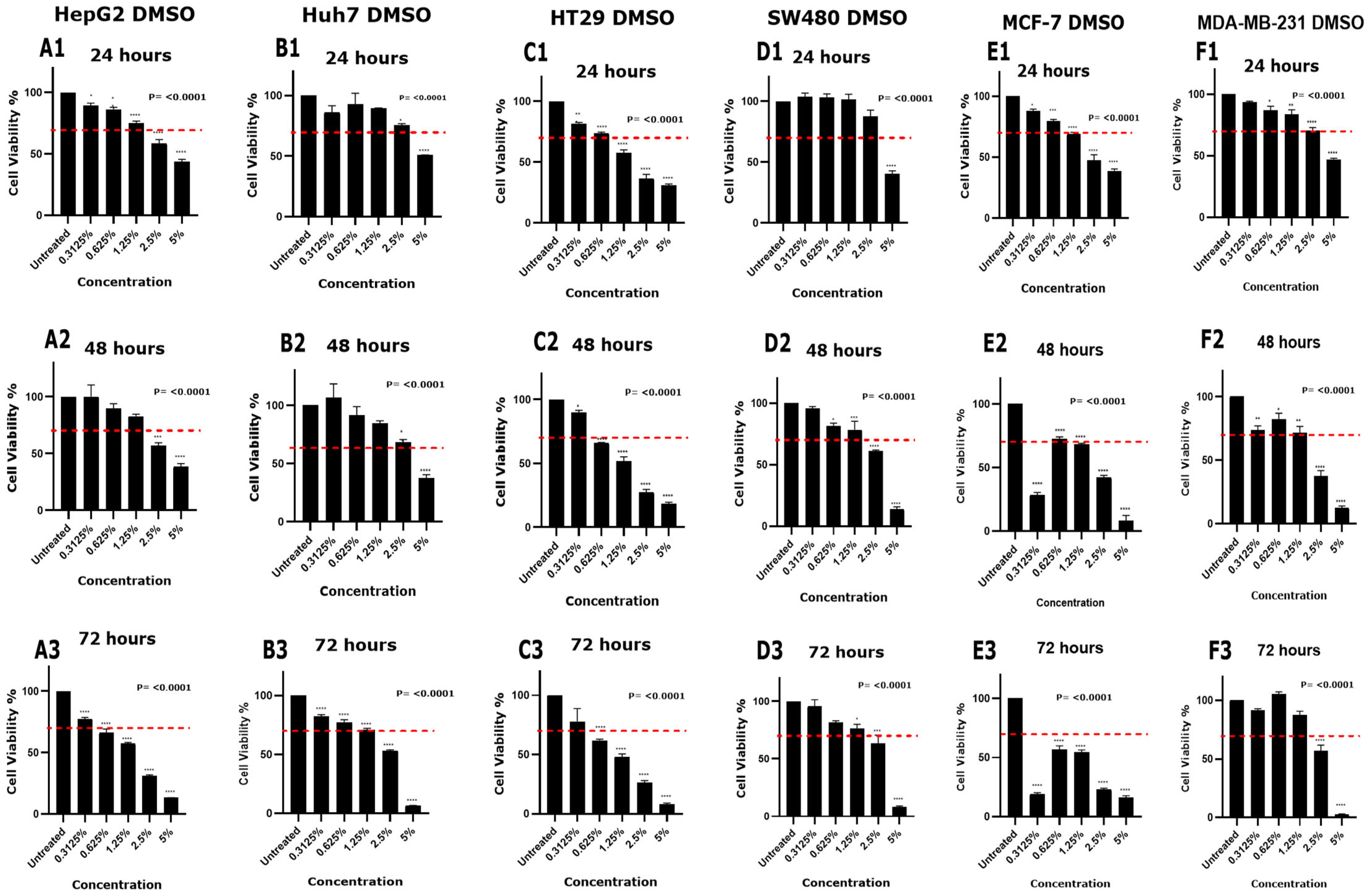
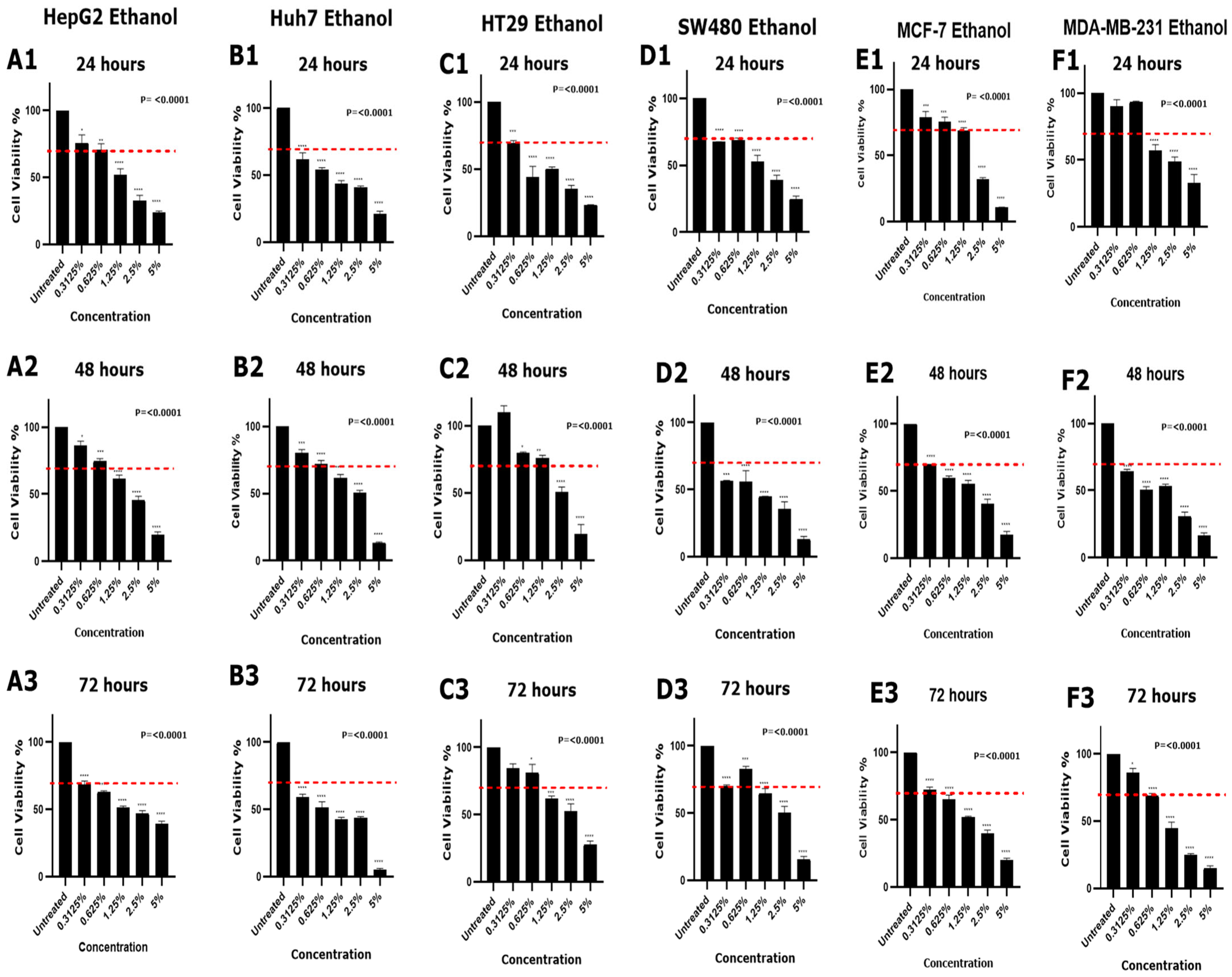
3.2. Effect of DMSO and Ethanol on Cell Viability in Various Cell Lines
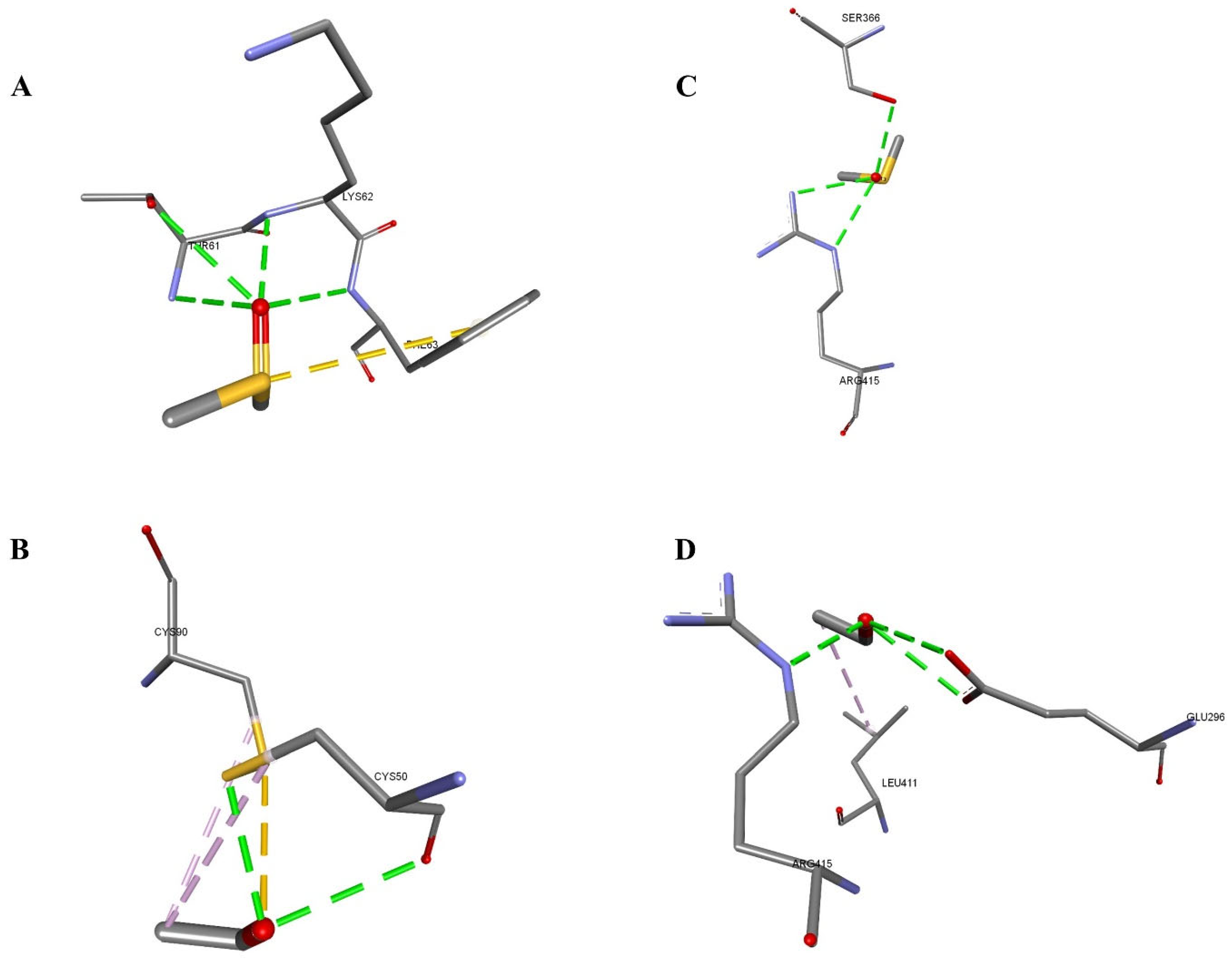
3.3. In Silico Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Å | Angstrom (unit of length = 10−10 m) |
| ABCB1 | ATP-Binding Cassette Subfamily B Member 1 |
| BAX | BCL2 Associated X Protein |
| CRC | Colorectal Cancer |
| CYP2E1 | Cytochrome P450 Family 2 Subfamily E Member 1 |
| DMSO | Dimethyl Sulfoxide |
| DNA | Deoxyribonucleic Acid |
| HCC | Hepatocellular Carcinoma |
| HepG2 | Human Hepatocellular Carcinoma Cell Line |
| HT29 | Human Colorectal Adenocarcinoma Cell Line |
| Huh7 | Human Hepatoma Cell Line |
| iGemDock | Integrated Genetic Evolutionary Molecular Docking |
| LXRα | Liver X Receptor alpha |
| MCF-7 | Michigan Cancer Foundation-7 (Breast cancer cell line) |
| MDA-MB-231 | M.D. Anderson-Metastatic Breast 231 (Breast cancer cell line) |
| MTT | 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide |
| NR1H3 | Nuclear Receptor Subfamily 1 Group H Member 3 |
| PDB | Protein Data Bank |
| PDB ID | Protein Data Bank Identifier |
| P-gp | P-glycoprotein |
| PLA2G4A | Phospholipase A2 Group IVA |
| PyMol | Python Molecular Graphics Tool |
| SD | Standard Deviation |
| SW480 | Human Colon Adenocarcinoma Cell Line |
| v2.1 | Version 2.1 |
| vDW or vdW | Van der Waals |
References
- Gu, Y.; Yang, R.; Zhang, Y.; Guo, M.; Takehiro, K.; Zhan, M.; Yang, L.; Wang, H. Molecular mechanisms and therapeutic strategies in overcoming chemotherapy resistance in cancer. Mol. Biomed. 2025, 6, 2. [Google Scholar] [CrossRef]
- Kitaeva, K.V.; Rutland, C.S.; Rizvanov, A.A.; Solovyeva, V.V. Cell Culture Based in vitro Test Systems for Anticancer Drug Screening. Front. Bioeng. Biotechnol. 2020, 8, 322. [Google Scholar] [CrossRef]
- Niu, N.; Wang, L. In vitro human cell line models to predict clinical response to anticancer drugs. Pharmacogenomics 2015, 16, 273–285. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Buranaamnuay, K. The MTT assay application to measure the viability of spermatozoa: A variety of the assay protocols. Open Vet. J. 2021, 11, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, M.; Turnbull, T.; Sebastian, S.; Kempson, I. The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis. Int. J. Mol. Sci. 2021, 22, 12827. [Google Scholar] [CrossRef]
- Sylvester, P.W. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods Mol. Biol. 2011, 716, 157–168. [Google Scholar] [CrossRef]
- Petiti, J.; Revel, L.; Divieto, C. Optimizing resazurin-based viability assays for P-MSC/TER308 cell line to enhance results reliability. BMC Res. Notes 2025, 18, 228. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, Q.; Zhai, X.; Liu, L.; Hong, Y. Screening and validation of the optimal panel of reference genes in colonic epithelium and relative cancer cell lines. Sci. Rep. 2023, 13, 17777. [Google Scholar] [CrossRef] [PubMed]
- Boix-Montesinos, P.; Soriano-Teruel, P.M.; Arminan, A.; Orzaez, M.; Vicent, M.J. The past, present, and future of breast cancer models for nanomedicine development. Adv. Drug Deliv. Rev. 2021, 173, 306–330. [Google Scholar] [CrossRef] [PubMed]
- Arzumanian, V.; Pyatnitskiy, M.; Poverennaya, E. Comparative Transcriptomic Analysis of Three Common Liver Cell Lines. Int. J. Mol. Sci. 2023, 24, 8791. [Google Scholar] [CrossRef]
- Tuncer, S.; Gurbanov, R.; Sheraj, I.; Solel, E.; Esenturk, O.; Banerjee, S. Low dose dimethyl sulfoxide driven gross molecular changes have the potential to interfere with various cellular processes. Sci. Rep. 2018, 8, 14828. [Google Scholar] [CrossRef]
- Verheijen, M.; Lienhard, M.; Schrooders, Y.; Clayton, O.; Nudischer, R.; Boerno, S.; Timmermann, B.; Selevsek, N.; Schlapbach, R.; Gmuender, H.; et al. DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Sci. Rep. 2019, 9, 4641. [Google Scholar] [CrossRef]
- Moskot, M.; Jakobkiewicz-Banecka, J.; Kloska, A.; Piotrowska, E.; Narajczyk, M.; Gabig-Ciminska, M. The Role of Dimethyl Sulfoxide (DMSO) in Gene Expression Modulation and Glycosaminoglycan Metabolism in Lysosomal Storage Disorders on an Example of Mucopolysaccharidosis. Int. J. Mol. Sci. 2019, 20, 304. [Google Scholar] [CrossRef]
- Smith, M.G.; Des Etages, S.G.; Snyder, M. Microbial synergy via an ethanol-triggered pathway. Mol. Cell. Biol. 2004, 24, 3874–3884. [Google Scholar] [CrossRef] [PubMed]
- Casanas-Sanchez, V.; Perez, J.A.; Quinto-Alemany, D.; Diaz, M. Sub-toxic Ethanol Exposure Modulates Gene Expression and Enzyme Activity of Antioxidant Systems to Provide Neuroprotection in Hippocampal HT22 Cells. Front. Physiol. 2016, 7, 312. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, J.; Yang, C.; Wang, Y.; Yang, Y.; Qiu, P.; Xie, W.; Zhang, S.; Lu, T. Effects of Solvent Dimethyl Sulfoxide Invites a Rethink of Its Application in Amyloid Beta Cytotoxicity. Int. J. Toxicol. 2025, 44, 10915818251338235. [Google Scholar] [CrossRef]
- Timm, M.; Saaby, L.; Moesby, L.; Hansen, E.W. Considerations regarding use of solvents in in vitro cell based assays. Cytotechnology 2013, 65, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Gao, J.; Guo, J.; Bai, L.; Marshall, C.; Cai, Z.; Wang, L.; Xiao, M. Dimethyl sulfoxide damages mitochondrial integrity and membrane potential in cultured astrocytes. PLoS ONE 2014, 9, e107447. [Google Scholar] [CrossRef]
- Tamagawa, S.; Sakai, D.; Schol, J.; Sako, K.; Nakamura, Y.; Matsushita, E.; Warita, T.; Hazuki, S.; Nojiri, H.; Sato, M.; et al. N-acetylcysteine attenuates oxidative stress-mediated cell viability loss induced by dimethyl sulfoxide in cryopreservation of human nucleus pulposus cells: A potential solution for mass production. JOR Spine 2022, 5, e1223. [Google Scholar] [CrossRef]
- Toth, M.E.; Vigh, L.; Santha, M. Alcohol stress, membranes, and chaperones. Cell Stress Chaperones 2014, 19, 299–309. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Alfarsi, A.; Nguyen, H.T.; Davidson, G.; Lloyd, N.D.R.; Kumar, A. Metabolic disruptions induced by low concentrations of DMSO in RTgill-W1 fish cells: The importance of solvent controls in in vitro studies. Aquat. Toxicol. 2025, 283, 107354. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.T.; Nguyen, H.T.L.; Truong, K.D. Comparative cytotoxic effects of methanol, ethanol and DMSO on human cancer cell lines. Biomed. Res. Ther. 2020, 7, 3855–3859. [Google Scholar] [CrossRef]
- Demir, E.A.; Demir, S.; Aliyazıcıoğlu, Y. In vitro Cytotoxic Effect of Ethanol and Dimethyl Sulfoxide on Various Human Cell Lines. J. Agric. Nat. 2020, 23, 1119–1124. [Google Scholar] [CrossRef]
- Koc, A.; Karabay, A.Z.; Özkan, T.; Buyukbingol, Z.; Aktan, F. Time and Concentration Dependent Effects of Different Solvents on Proliferation of K562, HL60, HCT-116 and H929 Cell Lines. J. Res. Pharm. 2022, 26, 494–501. [Google Scholar] [CrossRef]
- Szlasa, W.; Zendran, I.; Zalesinska, A.; Tarek, M.; Kulbacka, J. Lipid composition of the cancer cell membrane. J. Bioenerg. Biomembr. 2020, 52, 321–342. [Google Scholar] [CrossRef]
- Cherkaoui, S.; Durot, S.; Bradley, J.; Critchlow, S.; Dubuis, S.; Masiero, M.M.; Wegmann, R.; Snijder, B.; Othman, A.; Bendtsen, C.; et al. A functional analysis of 180 cancer cell lines reveals conserved intrinsic metabolic programs. Mol. Syst. Biol. 2022, 18, e11033. [Google Scholar] [CrossRef]
- McKim, J.M., Jr. Building a tiered approach to in vitro predictive toxicity screening: A focus on assays with in vivo relevance. Comb. Chem. High Throughput Screen. 2010, 13, 188–206. [Google Scholar] [CrossRef]
- Gruber, S.; Nickel, A. Toxic or not toxic? The specifications of the standard ISO 10993-5 are not explicit enough to yield comparable results in the cytotoxicity assessment of an identical medical device. Front. Med. Technol. 2023, 5, 1195529. [Google Scholar] [CrossRef]
- ISO 10993-5:2009; Biological Evaluation of Medical Devices; Part 5: Tests for In Vitro Cytotoxicity. ISO, International Organization for Standardization: Geneva, Switzerland, 2009.
- van Tonder, A.; Joubert, A.M.; Cromarty, A.D. Limitations of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay when compared to three commonly used cell enumeration assays. BMC Res. Notes 2015, 8, 47. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Zhou, B.; Ho, S.S.; Greer, S.U.; Spies, N.; Bell, J.M.; Zhang, X.; Zhu, X.; Arthur, J.G.; Byeon, S.; Pattni, R.; et al. Haplotype-resolved and integrated genome analysis of the cancer cell line HepG2. Nucleic Acids Res. 2019, 47, 3846–3861. [Google Scholar] [CrossRef]
- Kawamoto, M.; Yamaji, T.; Saito, K.; Shirasago, Y.; Satomura, K.; Endo, T.; Fukasawa, M.; Hanada, K.; Osada, N. Identification of Characteristic Genomic Markers in Human Hepatoma HuH-7 and Huh7.5.1-8 Cell Lines. Front. Genet. 2020, 11, 546106. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Nitisa, D.; Pirsko, V.; Cakstina, I. Selecting suitable reference genes for qPCR normalization: A comprehensive analysis in MCF-7 breast cancer cell line. BMC Mol. Cell Biol. 2020, 21, 68. [Google Scholar] [CrossRef] [PubMed]
- Amaro, A.; Angelini, G.; Mirisola, V.; Esposito, A.I.; Reverberi, D.; Matis, S.; Maffei, M.; Giaretti, W.; Viale, M.; Gangemi, R.; et al. A highly invasive subpopulation of MDA-MB-231 breast cancer cells shows accelerated growth, differential chemoresistance, features of apocrine tumors and reduced tumorigenicity in vivo. Oncotarget 2016, 7, 68803–68820. [Google Scholar] [CrossRef]
- Hewitt, R.E.; McMarlin, A.; Kleiner, D.; Wersto, R.; Martin, P.; Tsokos, M.; Stamp, G.W.; Stetler-Stevenson, W.G. Validation of a model of colon cancer progression. J. Pathol. 2000, 192, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Gietema, J.A.; Shida, S.; Selvakumaran, M.; Fonrose, X.; Haas, N.B.; Testa, J.; O’Dwyer, P.J. In vitro hypoxia-conditioned colon cancer cell lines derived from HCT116 and HT29 exhibit altered apoptosis susceptibility and a more angiogenic profile in vivo. Br. J. Cancer 2005, 93, 1356–1363. [Google Scholar] [CrossRef]
- Punyamurtula, U.; Brown, T.W.; Zhang, S.; George, A.; El-Deiry, W.S. Cancer cell seeding density as a mechanism of chemotherapy resistance: A novel cancer cell density index based on IC50-Seeding Density Slope (ISDS) to assess chemosensitivity. Am. J. Cancer Res. 2023, 13, 5914–5933. [Google Scholar]
- Niepel, M.; Hafner, M.; Chung, M.; Sorger, P.K. Measuring Cancer Drug Sensitivity and Resistance in Cultured Cells. Curr. Protoc. Chem. Biol. 2017, 9, 55–74. [Google Scholar] [CrossRef]
- Banic, B.; Nipic, D.; Suput, D.; Milisav, I. DMSO modulates the pathway of apoptosis triggering. Cell. Mol. Biol. Lett. 2011, 16, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Koiri, R.K.; Trigun, S.K. Dimethyl sulfoxide activates tumor necrosis factoralpha-p53 mediated apoptosis and down regulates D-fructose-6-phosphate-2-kinase and lactate dehydrogenase-5 in Dalton’s lymphoma in vivo. Leuk. Res. 2011, 35, 950–956. [Google Scholar] [CrossRef]
- Pal, R.; Mamidi, M.K.; Das, A.K.; Bhonde, R. Diverse effects of dimethyl sulfoxide (DMSO) on the differentiation potential of human embryonic stem cells. Arch. Toxicol. 2012, 86, 651–661. [Google Scholar] [CrossRef]
- Sangweni, N.F.; Dludla, P.V.; Chellan, N.; Mabasa, L.; Sharma, J.R.; Johnson, R. The Implication of Low Dose Dimethyl Sulfoxide on Mitochondrial Function and Oxidative Damage in Cultured Cardiac and Cancer Cells. Molecules 2021, 26, 7305. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, P.C.; Dehghani, A.C.; Theriau, C.F.; Connor, M.K.; Perry, C.G.R. Synergistic activation of mitochondrial metabolism and the glutathione redox couple protects HepG2 hepatocarcinoma cells from palmitoylcarnitine-induced stress. Am. J. Physiol. Cell Physiol. 2019, 317, C1324–C1329. [Google Scholar] [CrossRef]
- Bohusne Barta, B.; Simon, A.; Nagy, L.; Danko, T.; Raffay, R.E.; Petovari, G.; Zsiros, V.; Sebestyen, A.; Sipos, F.; Muzes, G. Survival of HT29 cancer cells is influenced by hepatocyte growth factor receptor inhibition through modulation of self-DNA-triggered TLR9-dependent autophagy response. PLoS ONE 2022, 17, e0268217. [Google Scholar] [CrossRef]
- Donato, M.T.; Tolosa, L.; Gomez-Lechon, M.J. Culture and Functional Characterization of Human Hepatoma HepG2 Cells. Methods Mol. Biol. 2015, 1250, 77–93. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Gill, E.E.; Coombs, M.R.P.; Falsafi, R.; Hancock, R.E.W.; Hoskin, D.W. MDA-MB-231 Breast Cancer Cells Resistant to Pleurocidin-Family Lytic Peptides Are Chemosensitive and Exhibit Reduced Tumor-Forming Capacity. Biomolecules 2020, 10, 1220. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Smirnova, O.A.; Ivanova, O.N.; Masalova, O.V.; Kochetkov, S.N.; Isaguliants, M.G. Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells. PLoS ONE 2011, 6, e24957. [Google Scholar] [CrossRef] [PubMed]
- Rohwedder, A.; Zhang, Q.; Rudge, S.A.; Wakelam, M.J. Lipid droplet formation in response to oleic acid in Huh-7 cells is mediated by the fatty acid receptor FFAR4. J. Cell Sci. 2014, 127, 3104–3115. [Google Scholar] [CrossRef]
- Miyamoto, M.; Murphy, T.H.; Schnaar, R.L.; Coyle, J.T. Antioxidants protect against glutamate-induced cytotoxicity in a neuronal cell line. J. Pharmacol. Exp. Ther. 1989, 250, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, C.M.; Baldivia, D.D.S.; de Castro, D.T.H.; Carvalho, J.T.G.; Oliveira, A.S.; da Rocha, P.D.S.; Campos, J.F.; Balogun, S.O.; de Oliveira, C.F.R.; da Silva, D.B.; et al. Chemical Composition, Antioxidant, and Cytotoxic Effects of Senna rugosa Leaf and Root Extracts on Human Leukemia Cell Lines. Pharmaceuticals 2024, 17, 974. [Google Scholar] [CrossRef] [PubMed]
- Worley, S.L.; Vaughn, B.J.; Terry, A.I.; Gardiner, C.S.; DeKrey, G.K. Time- and dose-dependent effects of ethanol on mouse embryonic stem cells. Reprod. Toxicol. 2015, 57, 157–164. [Google Scholar] [CrossRef]
- Higuchi, H.; Adachi, M.; Miura, S.; Gores, G.J.; Ishii, H. The mitochondrial permeability transition contributes to acute ethanol-induced apoptosis in rat hepatocytes. Hepatology 2001, 34, 320–328. [Google Scholar] [CrossRef]
- Rodriguez, F.D.; Simonsson, P.; Gustavsson, L.; Alling, C. Mechanisms of adaptation to the effects of ethanol on activation of phospholipase C in NG 108-15 cells. Neuropharmacology 1992, 31, 1157–1164. [Google Scholar] [CrossRef]
- Cao, H.; Wei, D.; Yang, Y.; Shang, Y.; Li, G.; Zhou, Y.; Ma, Q.; Xu, Y. Systems-level understanding of ethanol-induced stresses and adaptation in E. coli. Sci. Rep. 2017, 7, 44150. [Google Scholar] [CrossRef]
- Gerets, H.H.; Tilmant, K.; Gerin, B.; Chanteux, H.; Depelchin, B.O.; Dhalluin, S.; Atienzar, F.A. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol. Toxicol. 2012, 28, 69–87. [Google Scholar] [CrossRef]
- Hoang, B.X.; Han, B.O.; Fang, W.H.; Tran, H.D.; Hoang, C.; Shaw, D.G.; Nguyen, T.Q. The Rationality of Implementation of Dimethyl Sulfoxide as Differentiation-inducing Agent in Cancer Therapy. Cancer Diagn. Progn. 2023, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, F.; Kinne, R.K. Ethanol treatment of hepatocellular carcinoma: High potentials of low concentrations. Cancer Biol. Ther. 2004, 3, 430–433. [Google Scholar] [CrossRef]
- Kar, N.; Gupta, D.; Bellare, J. Ethanol affects fibroblast behavior differentially at low and high doses: A comprehensive, dose-response evaluation. Toxicol. Rep. 2021, 8, 1054–1066. [Google Scholar] [CrossRef]

| DMSO Concentration | Cytotoxicity Effect (>30% Reduction in Cell Viability) * | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HepG2 | Huh7 | HT29 | SW480 | MCF-7 | MDA-MB-231 | |||||||||||||
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |
| 0.3125% | No | No | No | No | No | No | No | No | No | No | No | No | No | Yes | Yes | No | No | No |
| 0.625% | No | No | Yes | No | No | No | No | Yes | Yes | No | No | No | No | No | Yes | No | No | No |
| 1.25% | No | No | Yes | No | No | No | Yes | Yes | Yes | No | No | No | Yes | Yes | Yes | No | No | No |
| 2.5% | Yes | Yes | Yes | No | No | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| 5% | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Ethanol Concentration | Cytotoxicity Effect (>30% Reduction in Cell Viability) * | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HepG2 | Huh7 | HT29 | SW480 | MCF-7 | MDA-MB-231 | |||||||||||||
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |
| 0.3125% | No | No | Yes | Yes | No | Yes | Yes | No | No | Yes | Yes | No | No | Yes | No | No | Yes | No |
| 0.625% | No | No | Yes | Yes | No | Yes | Yes | No | No | Yes | Yes | No | No | Yes | Yes | No | Yes | Yes |
| 1.25% | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 2.5% | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 5% | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Protein–Solvent | Energy (kcal/mol) | vdW (kcal/mol) | Interacting Residues (Protein-Ligand) | Distance (Å) | Bond Type |
|---|---|---|---|---|---|
| 3E6I–DMSO | −32.443 | −25.443 | A:LEU463:N–Z:PRE999:O | 2.65103 | Conventional H-Bond |
| A:VAL464:N–Z:PRE999:O | 3.04291 | Conventional H-Bond | |||
| 3E6I–Ethanol | −37.3332 | −20.248 | A:ARG100:NH1–Z:PRE999:O | 2.61927 | Conventional H-Bond |
| A:ARG435:NH1–Z:PRE999:O | 2.65332 | Conventional H-Bond | |||
| Z:PRE999:O–A:ILE114:O | 2.86426 | Conventional H-Bond | |||
| Z:PRE999:O–A:ILE115:O | 3.13713 | Conventional H-Bond | |||
| Z:PRE999:C–A:ILE114 | 4.96809 | Alkyl | |||
| Z:PRE999:C–A:ARG435 | 3.64147 | Alkyl | |||
| A:TRP122–Z:PRE999:C | 4.86274 | Pi-Alkyl | |||
| A:TRP122–Z:PRE999:C | 5.38205 | Pi-Alkyl | |||
| 6QEX–DMSO | −36.256 | −20.7804 | A:LYS1076:N–Z:PRE999:O | 2.84994 | Conventional H-Bond |
| A:SER1077:N–Z:PRE999:O | 2.8904 | Conventional H-Bond | |||
| A:SER1077:OG–Z:PRE999:O | 3.10488 | Conventional H-Bond | |||
| A:THR1078:N–Z:PRE999:O | 2.94488 | Conventional H-Bond | |||
| A:THR1078:OG1–Z:PRE999:O | 2.70126 | Conventional H-Bond | |||
| 6QEX–Ethanol | −35.8352 | −18.9044 | A:GLY430:N–Z:PRE999:O | 3.08366 | Conventional H-Bond |
| A:CYS431:N–Z:PRE999:O | 2.9703 | Conventional H-Bond | |||
| A:GLY432:N–Z:PRE999:O | 2.78409 | Conventional H-Bond | |||
| A:LYS433:NZ–Z:PRE999:O | 2.89953 | Conventional H-Bond | |||
| Z:PRE999:O–A:ASN428:O | 3.10461 | Conventional H-Bond |
| Protein–Solvent | Energy | vDW | Interacting Residues (Protein-Ligand) | Distance (Å) | Bond Type |
|---|---|---|---|---|---|
| 1DB4-DMSO | −33.14 | −20.61 | A:THR61:N-Z:PRE999:O | 3.05995 | Conventional Hydrogen Bond |
| A:THR61:OG1-Z:PRE999:O | 3.19181 | Conventional Hydrogen Bond | |||
| A:LYS62:N-Z:PRE999:O | 2.62457 | Conventional Hydrogen Bond | |||
| A:PHE63:N-Z:PRE999:O | 3.10118 | Conventional Hydrogen Bond | |||
| Z:PRE999:S-A:PHE63 | 4.61255 | Pi-Sulfur | |||
| 1DB4-Ethanol | −30.52 | −17.0 | Z:PRE999:O-A:CYS50:O | 3.09723 | Conventional Hydrogen Bond |
| Z:PRE999:O-A:CYS50:SG | 2.71761 | Conventional Hydrogen Bond | |||
| A:CYS90:SG-Z:PRE999:O | 3.24338 | Sulfur-X | |||
| Z:PRE999:C-A:CYS50 | 4.47882 | Alkyl | |||
| Z:PRE999:C-A:CYS90 | 4.41033 | Alkyl | |||
| 1UHL-DMSO | −37.78 | −21.95 | B:SER366:OG-Z:PRE999:O | 2.88224 | Conventional Hydrogen Bond |
| B:ARG415:NE-Z:PRE999:O | 2.97604 | Conventional Hydrogen Bond | |||
| B:ARG415:NH2-Z:PRE999:O | 2.82177 | Conventional Hydrogen Bond | |||
| 1UHL-Ethanol | −36.06 | −15.27 | B:ARG415:NE-Z:PRE999:O | 2.71453 | Conventional Hydrogen Bond |
| Z:PRE999:O-B:GLU296:OE1 | 3.11114 | Conventional Hydrogen Bond | |||
| Z:PRE999:O-B:GLU296:OE2 | 3.1035 | Conventional Hydrogen Bond | |||
| Z:PRE999:C-B:LEU411 | 4.09698 | Alkyl |
| Protein–Solvent | Energy | vDW | Interacting Residues (Protein-Ligand) | Distance (Å) | Bond Type |
|---|---|---|---|---|---|
| 3KJF-DMSO | −38.13 | −26.53 | B:LYS260:N-Z:PRE999:O | 3.13923 | Conventional Hydrogen Bond |
| Z:PRE999:O-A:ASP169:O | 3.12752 | Conventional Hydrogen Bond | |||
| Z:PRE999:O-B:LYS260:O | 2.59778 | Conventional Hydrogen Bond | |||
| B:TYR203-Z:PRE999:C | 4.93447 | Pi-Alkyl | |||
| B:TRP206-Z:PRE999:C | 4.60801 | Pi-Alkyl | |||
| B:TRP206-Z:PRE999:C | 5.45039 | Pi-Alkyl | |||
| 3KJF-Ethanol | −37.01 | −21.94 | A:ARG64:NH2-Z:PRE999:O | 2.60446 | Conventional Hydrogen Bond |
| B:ARG207:NE-Z:PRE999:O | 3.1156 | Conventional Hydrogen Bond | |||
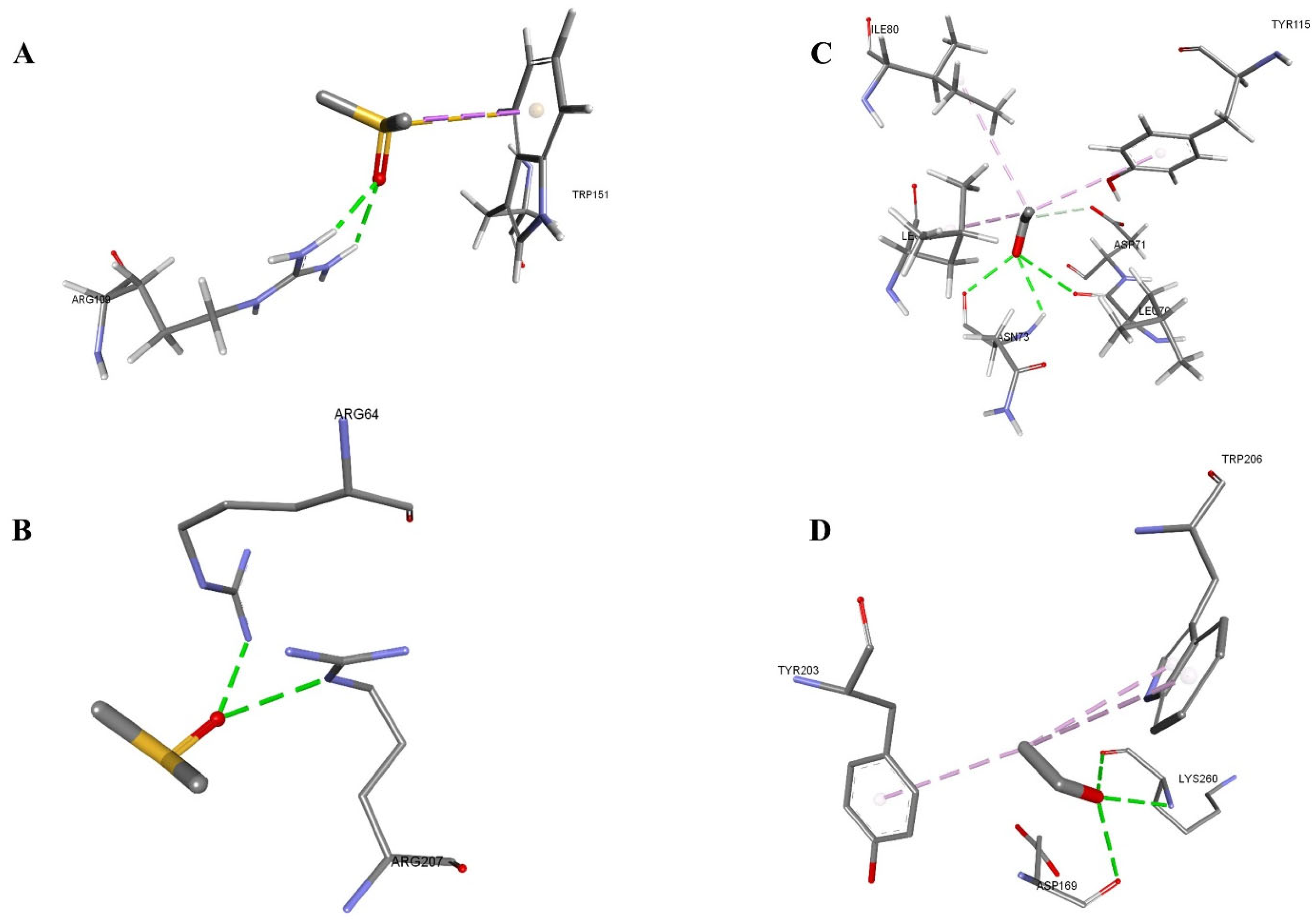
| 1F16-DMSO | −34.49 | −27.49 | A:ARG109:HH12-Z:PRE999:O | 2.19023 | Conventional Hydrogen Bond |
| A:ARG109:HH22-Z:PRE999:O | 2.25436 | Conventional Hydrogen Bond | |||
| Z:PRE999:C-A:TRP151 | 3.66643 | Pi-Sigma | |||
| Z:PRE999:S-A:TRP151 | 3.84053 | Pi-Sulfur | |||
| 1F16-Ethanol | −33.12 | −22.77 | A:ASN73:H-Z:PRE999:O | 2.66065 | Conventional Hydrogen Bond |
| Z:PRE999:O-A:LEU70:O | 3.0568 | Conventional Hydrogen Bond | |||
| Z:PRE999:O-A:ASN73:O | 2.52568 | Conventional Hydrogen Bond | |||
| Z:PRE999:C-A:ASP71:OD1 | 3.70876 | Carbon Hydrogen Bond | |||
| Z:PRE999:C-A:LEU76 | 3.84647 | Alkyl | |||
| Z:PRE999:C-A:ILE80 | 5.19701 | Alkyl | |||
| A:TYR115-Z:PRE999:C | 5.24314 | Pi-Alkyl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asiri, A.; Tasleem, M.; Al Said, M.; Asiri, A.; Al Qarni, A.A.; Bakillah, A. Optimizing Cell Density and Unveiling Cytotoxic Profiles of DMSO and Ethanol in Six Cancer Cell Lines: Experimental and In Silico Insights. Methods Protoc. 2025, 8, 93. https://doi.org/10.3390/mps8040093
Asiri A, Tasleem M, Al Said M, Asiri A, Al Qarni AA, Bakillah A. Optimizing Cell Density and Unveiling Cytotoxic Profiles of DMSO and Ethanol in Six Cancer Cell Lines: Experimental and In Silico Insights. Methods and Protocols. 2025; 8(4):93. https://doi.org/10.3390/mps8040093
Chicago/Turabian StyleAsiri, Abutaleb, Munazzah Tasleem, Muwadah Al Said, Abdulaziz Asiri, Ali Ahmed Al Qarni, and Ahmed Bakillah. 2025. "Optimizing Cell Density and Unveiling Cytotoxic Profiles of DMSO and Ethanol in Six Cancer Cell Lines: Experimental and In Silico Insights" Methods and Protocols 8, no. 4: 93. https://doi.org/10.3390/mps8040093
APA StyleAsiri, A., Tasleem, M., Al Said, M., Asiri, A., Al Qarni, A. A., & Bakillah, A. (2025). Optimizing Cell Density and Unveiling Cytotoxic Profiles of DMSO and Ethanol in Six Cancer Cell Lines: Experimental and In Silico Insights. Methods and Protocols, 8(4), 93. https://doi.org/10.3390/mps8040093








