Abstract
This protocol paper outlines a robust and reproducible framework for a 1H nuclear magnetic resonance (NMR) metabolomics analysis of rodent plasma, designed to facilitate preclinical biomarker discovery. The protocol details optimised steps for plasma collection in a preclinical rodent model, sample preparation, and NMR data acquisition using presaturation Carr–Purcell–Meiboom–Gill (PRESAT-CPMG) pulse sequences, ensuring high-quality spectral data and effective suppression of macromolecule signals. Comprehensive spectral processing and metabolite assignment are described, with guidance on multivariate and univariate statistical analyses to identify metabolic changes and potential biomarkers. The framework emphasises methodological rigour and reproducibility, enabling accurate quantification and interpretation of metabolites relevant to disease mechanisms or therapeutic interventions. By providing a standardised approach, this protocol supports longitudinal and translational studies, bridging findings from rodent models to clinical applications and advancing the reliability of metabolomics-based biomarker discovery in preclinical research.
1. Introduction
Metabolomics is rapidly becoming indispensable to translational research, particularly for animal models where biofluids such as plasma and urine enable non-invasive monitoring of systemic metabolic changes in response to genetic, dietary, or therapeutic interventions [1]. These biofluids provide real-time and time-integrated snapshots of metabolic status, making them highly valuable for longitudinal studies and bridging findings from preclinical models to clinical applications [2]. For example, plasma metabolomics in rodent models of obesity and diabetes consistently identifies disturbances in amino acid metabolism (e.g., alanine, glutamine) and energy metabolism intermediates (e.g., lactate, citrate) as hallmarks of metabolic dysfunction [3,4]. Critically, these metabolic pathways and biomarkers align closely with those observed in human non-communicable disease (NCD) cohorts, underscoring the translational relevance of rodent studies [1,5].
Amongst the analytical platforms used in metabolomics, nuclear magnetic resonance (NMR) spectroscopy is especially invaluable. NMR allows for the simultaneous detection and quantification of a broad spectrum of metabolites, whilst its non-destructive nature enables repeated analyses of the same sample, thereby offering an advantage for longitudinal sampling application [1,6,7]. The Zucker Diabetic Fatty (ZDF) rat is a well-validated preclinical model for bridging translational research in type 2 diabetes (T2D), particularly due to its recapitulation of human metabolic and pathological disease progression [8]. ZDF rats develop severe hyperglycaemia, insulin resistance, and obesity under high-fat diets, mirroring the multifactorial aetiology of human T2D [9]. Recent studies in rodent models highlight the utility of NMR-based metabolomics for capturing metabolic flexibility. For instance, non-fasted plasma sampling in Nile rats (Arvicanthis niloticus), a model for T2D, demonstrated stronger associations with glucose tolerance and lower replicate variance compared to fasted samples, suggesting practical advantages for metabolic phenotyping [10]. Such methodological insights enhance the reliability of plasma metabolomics in reflecting systemic metabolic states.
The integration of advanced analytical software is crucial to extracting meaningful biological insights from complex NMR datasets. Firstly, targeted quantification using tools like Chenomx NMR Suite enables a precise identification and concentration determination of metabolites in biofluids [7,11]. Furthermore, the application of multivariate statistical tools, including those implemented in SIMCA-P software (version 17), facilitate the recognition of metabolic patterns through techniques including principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) [3]. After performing univariate analysis and robust biomarker identification, a multiple comparisons analysis is often performed to determine the groups that differ significantly [12,13,14,15]. Interpreting metabolomics data in studies involving mixed diets (e.g., whole foods or complex meals) presents challenges due to overlapping metabolic signals from multiple nutrients, interactions between dietary components, and individual variability in metabolism [16]. This contrasts with controlled interventions using single nutrients, where metabolic responses are more directly attributable to specific compounds [17]. The Benjamini–Hochberg multiple testing correction helps address these complexities by reducing false positives while maintaining sensitivity to subtle diet-related metabolic changes [18,19].
In summary, researchers can achieve reproducible and biologically relevant results in rodent models by combining standardised NMR acquisition, untargeted metabolite quantification, and integrated multivariate/univariate statistical analysis. This manuscript aims to provide a detailed, step-by-step protocol for a reproducible NMR-based metabolomics analysis of plasma in rodent models, integrating Chenomx NMR Suite (version 9) quantification, SIMCA-P (version 17) multivariate analysis, SPSS (version 26) statistical validation, and multiple testing correction. This manuscript aims to establish reproducible metabolomic workflows in preclinical research by employing a diabetic and obese rodent model, thereby bridging methodological gaps and improving the applicability of findings to human NCD studies.
2. Experimental Design
Twenty-four male Zucker Diabetic Fatty (ZDF fa/fa) rats (10 weeks old) and six lean male Zucker (wild-type) rats (10 weeks old) were sourced from Vital River (Beijing, China) for the experiment. ZDF rats possess a missense mutation in the leptin receptor (Lepr) gene, resulting in impaired leptin signalling and the spontaneous onset of obesity, insulin resistance, and T2D, as established by prior studies [20,21]. Following a 2-week quarantine, rats underwent a 1-week acclimatization under standardised conditions controlled for room temperature of 19–24 °C, 40–60% humidity, a 12/12 h light/dark cycle, ad libitum access to distilled water, and a standard rodent maintenance diet (11% fat, 65% carbohydrate, and 24% protein, expressed as % of total energy). Diabetes in the animal was confirmed pre-intervention via fasting blood glucose levels >200 mg/dL, as per established thresholds for diabetic phenotypes in ZDF rats [22].
ZDF rats were randomised into four groups (n = 6 per group), namely the control (standard rodent maintenance diet), Diet A (normal human diet), Diet B (low-carbohydrate–high-fat diet), and standard treatment (metformin, 200 mg/kg body weight daily). Lean Zucker rats (n = 6) were divided into two groups (n = 3 per group) receiving Diet A or B. After 8 weeks of intervention, rats were euthanised via 5% isoflurane inhalation, followed by cardiac puncture. Blood samples were collected into lithium heparin tubes, centrifuged (1500× g, 10 min, 4 °C), and plasma stored at −80 °C for NMR analysis. As the primary focus of this manuscript is on the NMR metabolomics workflow, details of the animal experimentation are therefore only briefly described.
2.1. Sample Size Calculation
The sample size for this study was determined using a one-way analysis of variance (ANOVA) approach, as described by [23]. The calculations utilised the degrees of freedom (DF) associated with the within-subject error term, which is defined as
where
- N: total number of subjects;
- k: number of experimental groups (k = 6);
- n: number of subjects per group.
Rearranging the formula to solve for n results in
Steps:
- DF Range: A DF range of 10 (minimum) to 20 (maximum) was selected to ensure statistical robustness while balancing practical constraints.
- Per-Group Calculation:
- Minimum n (DF = 10):
- Maximum n (DF = 20):
- 3.
- Total Sample Size:
- Minimum N (n = 3):
- Maximum N (n = 4):
2.2. Materials
- Deuterium oxide (D2O) (Merck, Frankfurter Strasse 250, Darmstadt, Germany, Cat. no.: 113366).
- Potassium dihydrogen phosphate (KH2PO4); ≥99.0% purity, (Merck, Frankfurter Strasse 250, Darmstadt, Germany, Cat. no.: 104873).
- Sodium 3-(trimethylsilyl) propionic-2,2,3,3-d4 (TSP) (Sigma-Aldrich, St. Louis, MO, USA, Cat. no.: 269913).
- Imidazole (Sigma-Aldrich, St. Louis, MO, USA, Cat. no.: I2399).
- Sodium deuteroxide (NaOD) (Merck, Frankfurter Strasse 250, Darmstadt, Germany, Cat. no.: 372072).
- Several 3 kDa centrifugal filters, 0.5 mL (Merck, Frankfurter Strasse 250, Darmstadt, Germany, Cat. no.: UFC500396).
- Safe-Lock micro test tubes, 1.5 mL (Eppendorf, Hamburg, Germany, Cat. no.: 0030120086).
- A long-form NMR pipette, tip length 9 inch (Sigma-Aldrich, St. Louis, MO, USA, Cat. no.: Z255688).
- Several 5 mm 600 MHz NMR tubes, length 7 inch (Norell, Morganton, NC, USA, Cat. no.: 509-UP-7).
- A 250 mL solvent bottle.
- An NMR tube rack for 5 mm tube (Sigma-Aldrich, St. Louis, MO, USA, Cat. no.: Z118257).
- Deionised water (for filter pre-washing).
- Ice.
- Dry ice.
- Liquid nitrogen.
2.3. Equipment
- A mechanical pipette (Eppendorf, Hamburg, Germany).
- A long-form NMR pipette, tip length 9 inch (Sigma-Aldrich, St. Louis, MO, USA, Cat. no.: Z255688).
- A vortex mixer (IKA, Staufen, Baden-Württemberg, Germany).
- A magnetic stirrer (IKA, Staufen, Baden-Württemberg, Germany).
- A high-speed centrifuge (Kubota, Osaka, Japan).
- A −80 °C Biomedical Freezer (SANYO, Osaka, Japan).
- A liquid nitrogen tank (Chart Biomedical, GA, USA).
- A pH metre (Hanna Instruments, RI, USA).
- A 600 MHz NMR Spectrometer (JEOL, Tokyo, Japan).
2.4. Computational Tools
- Chenomx NMR Suite (Chenomx Inc., Edmonton, AB, Canada).
- SIMCA-P (Sartorius Stedim Data Analytics AB, Göttingen, Germany).
- SPSS (IBM, Chicago, IL, USA).
3. Procedure
3.1. Sample Collection
This sample handling procedure is based on the recommendations outlined in [24], with a specific focus on minimising the processing time for serum and plasma samples.
- Blood collection: Collect blood via a cardiac puncture into lithium heparin tubes immediately after euthanasia after the end of the experiment.
 CRITICAL STEP EDTA or citrate tubes are not used, as these might introduce strong signals in NMR spectra, which could obscure adjacent metabolites.
CRITICAL STEP EDTA or citrate tubes are not used, as these might introduce strong signals in NMR spectra, which could obscure adjacent metabolites. - Plasma isolation: Centrifuge blood at 1500× g for 10 min at 4 °C within 30 min of blood collection.
- Storage: Aliquot plasma into 1.5 mL tubes and temporarily place on dry ice or in a liquid nitrogen tank for temporary storage. Transfer samples to −80 °C within 2–4 h of plasma separation.
3.2. NMR Sample Preparation
This procedure is modified from the method described in [25] to suit this study’s specific requirements. A general overview of the sample preparation is shown (Figure 1).
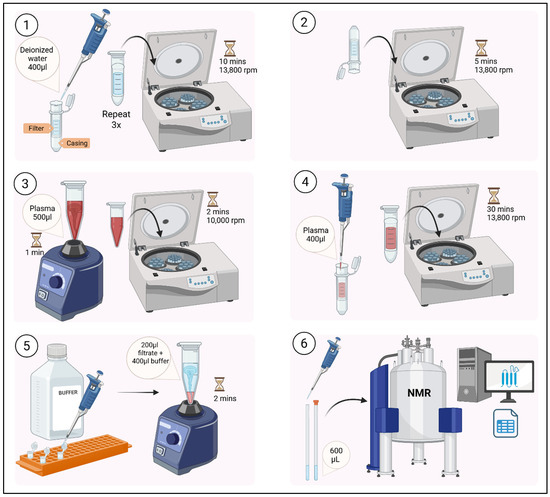
Figure 1.
Overview of sample preparation for metabolite profiling.
- Step 1: Thawing and preprocessing
- Thaw frozen plasma samples on ice on the day of NMR analysis.
- Vortex 500 μL plasma for 1 min.
- Centrifuge at 10,000 rpm for 2 min to pellet solid debris.
- Step 2: Macromolecule removal through ultrafiltration
- Pre-wash filters to remove glycerol: Rinse 3 kDa centrifugal filters with 400 µL deionised water. Centrifuge at 13,800 rpm for 10 min for each wash. Repeat this process twice.
- Remove residual water: Invert filters and centrifuge at 13,800 rpm for 5 min.
 CRITICAL STEP Ensure there are no traces of water inside the filter and casing. Use a pipette if necessary. Filter is now ready for usage.
CRITICAL STEP Ensure there are no traces of water inside the filter and casing. Use a pipette if necessary. Filter is now ready for usage. - Load 400 μL plasma supernatant thawed during Step 1 onto pre-washed filters.
- Centrifuge at 13,800 rpm for 30 min at room temperature to remove proteins/lipids.
Several studies have explored various methods for removing proteins from plasma and serum prior to NMR analysis, including solvent-based precipitation, ultrafiltration, and spectroscopic filtering [26,27,28]. Ultrafiltration is particularly advantageous for quantifying unbound, low-molecular-weight metabolites with minimal chemical artefacts. By applying a defined molecular weight cutoff, it reproducibly separates small metabolites from macromolecules like proteins and lipoproteins, improving spectral quality by reducing baseline distortion and signal overlap. Protein removal also mitigates matrix effects from differential metabolite binding, enhancing quantification consistency. Moreover, ultrafiltration avoids organic solvents, eliminating solvent-related artefacts. To address known limitations such as membrane interactions and glycerol contamination, pre-washing filters are recommended and incorporated into the current protocol [29]. While this protocol does not include a direct comparison of protein removal methods, ultrafiltration was selected based on support from the literature and practical benefits for workflow reproducibility and spectral clarity.
- Step 3: Buffer preparation and dilution
- Prepare a phosphate buffer (pH = 7.4) based on the volume required (e.g., 100 mL), with 1.232 g KH2PO4 in 100 mL D2O containing 100 mg TSP (0.1%) and 100 mg imidazole (0.1%). TSP functions as an internal standard, while imidazole functions as a pH indicator. The concentration of TSP in this buffer preparation is approximately 5.8 mM.
- Use a magnetic stirrer and stirrer bar to homogenise the buffer for 5 min.
- Add NaOD in small volumes ≈ 100 µL whilst homogenising the buffer using a magnetic stirrer for 5 min and checking the pH.
- Repeat step 3 to achieve pH = 7.4. Wrap the solvent bottle using aluminium foil and store at 4 °C until usage.
- Dilute filtrate with buffer in a 1:2 (v/v) ratio (e.g., 200 μL filtrate + 400 μL buffer) in a 1.5 mL tube.
- Vortex the tube for 2 min.
TSP is a commonly used internal standard in 1H NMR metabolomics, employed primarily for chemical shift referencing. In this study, we use TSP due to its sharp, isolated singlet at 0.00 ppm, which provides consistent and reliable chemical shift calibration across a broad range of biofluids. Its low pKa and lack of spectral overlap with endogenous metabolites make it well-suited for direct referencing, instrument calibration, and ensuring inter-batch consistency. TSP also remains a widely adopted reference in established metabolomics protocols, supporting cross-study comparability. However, we acknowledge the well-documented limitations of TSP in protein-rich biological matrices such as serum and plasma. As reported in [30], TSP binds strongly to human serum albumin, resulting in signal attenuation, peak broadening, and chemical shift variability, particularly in untreated samples. Because only the free (unbound) fraction of TSP contributes to a sharp, quantifiable resonance, this binding reduces its reliability as an internal standard for quantification. To address this, we combine ultrafiltration [31,32,33,34] with the use of TSP at elevated concentrations (≥3 mM), as suggested in [30]. Ultrafiltration effectively removes albumin and other proteins, thereby eliminating the bound TSP fraction and yielding a sharper, more reproducible reference peak. Simultaneously, the use of a higher TSP concentration maintains sufficient free TSP levels in the filtrate for accurate referencing.
- Step 4: Transfer to NMR tubes
- Pipette 600 μL of the diluted sample into a 5 mm NMR tube using a long-form NMR pipette.
 CRITICAL STEP Ensure that the NMR tubes are free of water which would interfere with the spectral acquisition. Use a tissue to make sure the external surface of the tube is completely dry.
CRITICAL STEP Ensure that the NMR tubes are free of water which would interfere with the spectral acquisition. Use a tissue to make sure the external surface of the tube is completely dry. - Seal tubes and store at 4 °C until spectral acquisition using an NMR spectrometer.
3.3. Quality Control
To ensure the NMR spectrometer remains in optimal condition throughout the experiment, pooled quality control (QC) samples are created by mixing aliquots from all study samples. These pooled QC samples are analysed qualitatively at the start, middle, and end of each day’s run. Any instrument drift, instability, or anomalies can be promptly identified by visually inspecting the QC spectra for consistent peak positions, line shapes, and signal-to-noise ratios (SNRs). This qualitative assessment helps verify the reliability of spectral acquisition across the entire experimental sequence.
3.4. NMR Acquisition
The following procedure aligns with the targeted metabolomics workflow described in [35] with several modifications:
- Instrument Setup:
- Use a 600 MHz JEOL JNM-ECZ600R NMR spectrometer (Jeol Ltd., Tokyo, Japan) maintained at 26 °C. Always use NMR tubes rated for the frequency of the spectrometer to ensure reliable, high-quality data and protect samples and the NMR instrument itself.
- Select the deuterium lock channel on the NMR spectrometer and lock onto the D2O signal.
- Perform shimming to optimise magnetic field homogeneity.
- After the sample is inserted and the temperature is stabilised, the spectrometer is locked onto the deuterium resonance (2H) of the D2O solvent. The automated gradient shimming routine is applied first; this function uses the deuterium lock signal to map the magnetic field profile across the sample volume and automatically adjusts shim coil currents to correct field inhomogeneities.
- The system uses a digital matrix shim set, which includes axial shim coils Z1 through Z6 corresponding to first- through sixth-order corrections along the z-axis. Specifically, Z1 compensates for linear gradients, Z2 for quadratic curvature, and Z3 to Z6 for higher-order magnetic field distortions. Following automatic shimming, the manual refinement of shim values (Z1–Z6) is carried out as needed. The adjustment sequence begins with Z1–Z3 to improve the 2H lock signal sharpness and continues with Z4–Z6 to correct subtle baseline distortions or peak asymmetries.
- Throughout the process, the stability of the lock signal and the shape of a reference peak (typically the water signal at ~4.7 ppm) are monitored. Shimming is considered complete when a narrow, symmetric reference peak and stable lock level are achieved.
- Apply the presaturation-Carr–Purcell–Meiboom–Gill (PRESAT-CPMG) pulse sequence to suppress water signals and protein resonances.
- 2.
- Spectral Acquisition Parameters:
- Spectral width: 12 ppm;
- Number of scans: 64;
- Total acquisition time: 26 min;
- Time domain points: 131,072 points;
- Flip angle: 90°;
- Pre-saturation frequency: set to water resonance (≈4.7 ppm in plasma);
- Acquisition time per scan: 17.47 s;
- Relaxation delay: 7 s;
- Repetition time: 24.47 s;
- Presaturation duration: 7 s;
- CPMG tau (τ) delay: 0.19291 milliseconds;
- Number of echoes (n): 125;
- Effective echo time (2τ): 0.38582 milliseconds;
- Total echo train duration (2nτ): 48.23 milliseconds.
A repetition time of approximately 25 s is used in the acquisition protocol, which corresponds to at least five times the longest longitudinal relaxation times (T1) reported in the literature for the metabolites of interest [36]. This repetition time is therefore considered sufficient to allow for a near-complete recovery of longitudinal magnetization and to ensure accurate quantitative analysis. It should be noted, however, that this assessment is based on T1 values reported in the literature, since no inversion recovery experiments are conducted to measure T1 directly in the current samples. The selection of the CPMG parameters for this study, specifically a tau (τ) delay of 0.19291 milliseconds, 125 echoes, an effective echo time (2τ) of 0.38582 milliseconds, and a total echo train duration (2nτ) of 48.23 milliseconds, is guided by a review of published 1H NMR metabolomics studies performed in rat plasma [37,38,39]. These parameters are chosen to achieve effective suppression of broad macromolecular signals, such as those from proteins and lipoproteins, while preserving sharp signals from low-molecular-weight metabolites.
Although the general approach to CPMG implementation in rodent metabolomics is conceptually consistent, the explicit reporting of critical sequence parameters such as the tau delay and number of echoes remains inconsistent. For example, ref. [37] applies a CPMG-based spin–echo sequence in rat plasma with a fixed echo train duration of 60 milliseconds, but it does not report the tau delay or the number of echoes used. Similarly, ref. [38] reports a total echo train duration of 320 milliseconds in rat plasma using a one-dimensional spin–echo sequence, but it also omits the tau delay and number of echoes. In addition, ref. [39] applies a total echo train duration of 64 milliseconds, but again, it does not specify the tau delay and number of echoes explicitly. By contrast, CPMG protocols in human plasma are often more thoroughly reported. For instance, ref. [40] employs a tau delay of 0.83 milliseconds with 128 echoes, resulting in a total echo train duration of 212.48 milliseconds. Similarly, both refs. [41,42] use a tau delay of 0.4 milliseconds and 80 echoes, producing a total duration of 64 milliseconds. In contrast, for the current study in rodent plasma, universally agreed-upon parameters are lacking, and many published studies do not explicitly report the tau delay, echo time, or number of echoes. As such, preliminary pilot experiments are performed to optimise parameters specific to our sample system. The final values, a tau delay of 0.19291 milliseconds, 125 echoes, and a total echo train duration of 48.23 milliseconds, are empirically derived.
- 3.
- Adaptability to Different Brands
While this protocol is optimised and validated using the JEOL JNM-ECZ600R, it is adaptable to other major NMR instrument brands such as Bruker and Agilent or Varian. The fundamental CPMG parameters, such as the tau delay, number of echoes, presaturation routines, and temperature stabilisation, are universally implemented across commercial NMR platforms, enabling the core methodology to be transferred or replicated with appropriate adjustments to instrument-specific pulse sequence syntax and acquisition commands. We recommend that users carefully review their hardware specifications, including achievable tau delay increments, maximum echo counts, and probe characteristics, as these may influence effective signal suppression and data quality. Additionally, operational procedures like shimming, water suppression, and system calibration can differ between platforms. To ensure compatibility and optimal performance, we strongly suggest liaising with your respective instrument vendor when adapting this protocol to a different system. To support reproducibility and cross-platform comparability, we emphasise the importance of conducting pilot calibration runs and fine-tuning key parameters as needed. Clear documentation and complete reporting of acquisition settings are crucial, especially when comparing datasets across laboratories or instrument brands.
3.5. Data Preprocessing
- Spectral Processing (Chenomx 9.0):
- Spectrum files are imported into Chenomx NMR Suite for post-acquisition processing.
- Prior to Fourier transformation, free induction decays (FIDs) are zero-filled to the next power of two if needed to improve digital resolution. The zero-filled FIDs are then Fourier-transformed to convert the time-domain signal into the frequency-domain spectrum (e.g., 131,072 points in this study), enhancing spectral resolution and enabling more precise peak identification.
- Apply an exponential line broadening (apodization) function of 0.5 Hz to enhance the SNR. Apply 1–2 Hz line broadening if the signal-to-noise ratio is critically poor and resolution loss is tolerable. Do note that broader peaks reduce integration accuracy for closely spaced resonances. Several points should be considered for selecting the degree of line broadening, as reviewed from other studies [43,44]:
- Setting an SNR threshold for quantitation: For example, a minimum SNR of 10:1 is often recommended for reliable peak detection and integration. This threshold can be determined using reference peaks such as TSP.
- Evaluating the risk of peak overlap: Greater line broadening is only justifiable when analyte peaks are well separated, as excessive broadening risks merging closely spaced resonances and introduces quantitation error.
- Consistent batch processing: The same apodization parameters should be applied to all spectra within a batch, as varying these settings can artificially skew SNR measurements and introduce operator-dependent variability.
- Perform autophasing to ensure consistent and accurate phase correction across all spectra.
- Apply baseline correction using the Whittaker spline algorithm across the full spectral range, excluding the water resonance region, to effectively eliminate baseline distortions, drifts, and offsets. This approach ensures a flat and stable baseline, which is critical for accurate peak integration, reliable quantification, and robust comparison between samples.
- Reference spectra to TSP (0 ppm) for chemical shift calibration and imidazole for pH monitoring.
- 2.
- Binning and Exclusion:
- Use intelligent binning (0.04 ppm) across 0.50–12.00 ppm.
- Exclude regions:
- Water: 4.77–4.86 ppm;
- Imidazole: 7.29–7.33 ppm and 8.24–8.32 ppm.
- Export binned data as a non-negative integral table for multivariate analysis.
3.6. Multivariate Data Analysis
- Data Preprocessing:
- Mean-centre and Pareto-scale variables (divide by √standard deviation).
- 2.
- Unsupervised Analysis (PCA):
- Perform PCA to assess clustering trends and outliers.
- Identify outliers using Hotelling’s T2 (95% confidence interval).
- 3.
- Supervised Analysis (PLS-DA):
- Conduct PLS-DA to maximise class separation.
- Validate models via CV-ANOVA (p < 0.05 for significance).
- Prioritise bins with VIP scores > 1.0 and visualise using S-plots.
While multivariate data analysis (MVDA) effectively identifies spectral regions of interest, it is critical to recognise that a single NMR spectral bin (e.g., 1.22 ppm) may encompass signals from multiple metabolites. Thus, MVDA alone cannot resolve structural ambiguity, necessitating targeted metabolite identification (e.g., via Chenomx spectral libraries) and quantification to confirm biological relevance and avoid misinterpretation.
3.7. Metabolite Identification and Quantification
The procedure below is a simplified and adapted version following the guidance outlined in the Chenomx NMR Suite V9 User Guide (Chenomx Inc., Edmonton, AB, Canada) [45].
- Library Matching (Chenomx 9.0):
- Compare spectral peaks to the 600 MHz Human Metabolome Database (HMDB) library with pH adjustment for untargeted metabolite identification.
- Match chemical shift, multiplicity, and coupling constants based on guidelines provided by Chenomx. Fit metabolites by overlaying library spectra onto experimental data, iteratively subtracting matched peaks. Apply deconvolution to the experimental spectrum for overlapping peaks using the reference peak’s observed line shape.
- Validate fits via visual inspection of residual spectra.
- 2.
- Quantification:
- Calculate relative concentrations using peak areas normalised to TSP.
- Export results as tables for downstream analysis.
3.8. Statistical Analysis
- Univariate Analysis and Multiple Testing CorrectionFor studies involving group comparisons (e.g., control vs. treated), metabolites showing significant differences are first identified using a one-way ANOVA (p < 0.05). Post hoc pairwise comparisons are performed via Tukey’s honest significant difference (HSD) test to control family-wise error rates. The Benjamini–Hochberg procedure is applied to account for multiple hypothesis tests across metabolites, controlling the false discovery rate (FDR) at 10%. Metabolites with FDR-adjusted p-values < 0.05 are retained as statistically significant.
- Workflow Integration
- ANOVA identifies metabolites with significant intergroup variation.
- Tukey-HSD pinpoints specific group differences (e.g., Diet A vs. Diet B).
- FDR correction ranks metabolites by significance and adjusts p-values to reduce false positives.
This hierarchical approach ensures rigorous statistical validation while maintaining interpretability in complex datasets.
3.9. Pathway Analysis
The procedure below is a simplified version for pathway analysis using MetaboAnalyst 6.0 [46]. This workflow is designed to guide users through the essential steps, from data preparation to result interpretation, ensuring reproducibility and clarity in metabolomics pathway analysis.
- Define a List of Features of Interest
Identify and extract a list of metabolites that are relevant to the experimental question.
- 2.
- Perform Pathway Analysis
Use statistical methods to determine which biological pathways are significantly over-represented in your list compared to what would be expected by chance. This typically involves comparing your feature list to curated pathway databases (such as Kyoto Encyclopedia of Genomes and Genes (KEGG)) and calculating impact scores and p-values.
- 3.
- Visualise and Interpret Results
Visualise the impacted pathways using enriched pathways, p-values, impact scores, and matched metabolites to help interpret the biological significance of your findings.
4. Expected Results
The application of this NMR-based metabolomics protocol is expected to yield robust and reproducible metabolic profiles from rodent biofluids, enabling the systematic identification of disease-related biomarkers and pathway alterations in rodent models of obesity and diabetes. A tentative list of the detected metabolites is shown (Table 1). For each metabolite, the corresponding HMDB ID, chemical shift values (in ppm), and signal multiplicities are provided. Multiplicity notation indicates splitting patterns in the NMR spectra, with s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), and dd (doublet of doublets).

Table 1.
Identified metabolites.
Figure 2 shows the overlay of six spectra representing each group in this study. Figure 3 illustrates the differences between samples processed with ultrafiltration and without ultrafiltration. Please note that this comparison is shown for illustrative purposes only and may not represent the full variability or quantitative differences across all samples. Multivariate analysis using SIMCA-P will reveal a clear clustering of experimental groups, such as diabetic versus control animals, with PCA typically explaining more than 70% of the variance, and outliers are set to be effectively identified and excluded using Hotelling’s T2. Supervised analysis by PLS-DA is anticipated to achieve strong separation between groups (R2Y > 0.8, Q2 > 0.5), with model validity supported by significant CV-ANOVA results (p < 0.05). The 3D PLS-DA score plot is shown (Figure 4). Spectral bins with variable importance in projection (VIP) scores greater than 1.0 will emerge as key discriminators between phenotypes.
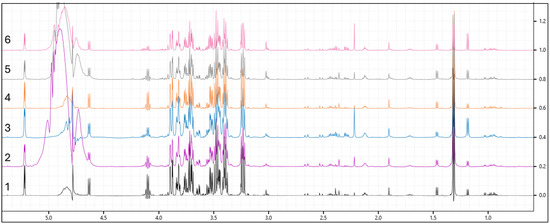
Figure 2.
Overlay of six spectra representing each group in this study. (1—control; 2—Diet A; 3—Diet B; 4—metformin; 5—lean with Diet A; and 6—lean with Diet B).
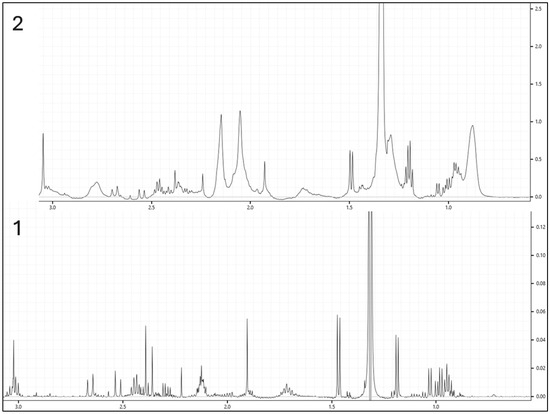
Figure 3.
Comparison of two samples processed with and without ultrafiltration. (1—processed with ultrafiltration; 2—processed without ultrafiltration).
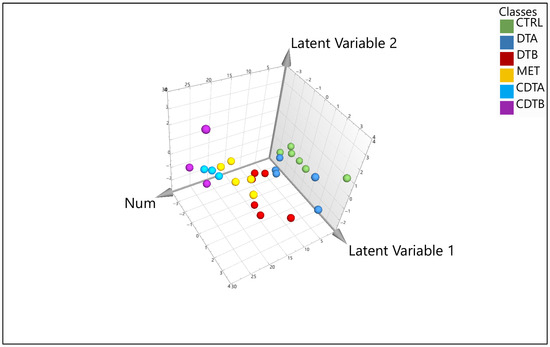
Figure 4.
Three-dimensional (3D) PLS-DA score plot for each sample. (Green colour—control; navy blue—Diet A; red—Diet B; yellow—metformin; light blue—lean with Diet A; and purple—lean with Diet B).
Metabolite identification and quantification using the Chenomx NMR Suite will enable a confident annotation of key metabolites, including branched-chain amino acids (leucine, valine), lactate, succinate, citrate, and glucose, with characteristic chemical shifts and spectral regions. The protocol’s preprocessing steps will ensure the exclusion of confounding signals, such as residual water and imidazole, resulting in clean and interpretable spectra. Statistical validation will reveal that numerous metabolites exhibit significant intergroup differences by an ANOVA, with post hoc Tukey-HSD tests pinpointing specific pairwise group differences. Following Benjamini–Hochberg correction for multiple tests (FDR = 10%), a subset of metabolites will retain statistical significance, reducing the risk of false positives. Biologically, the protocol is expected to capture hallmark metabolic changes associated with NCDs. Quality control samples will demonstrate high reproducibility, with coefficients of variation below 5% for major metabolites, and the use of intelligent binning will ensure consistent integration and minimal technical variability across replicates. Overall, these results will substantiate the protocol’s effectiveness for comprehensive, statistically rigorous metabolomic profiling in preclinical rodent studies. This protocol establishes a reproducible NMR-based metabolomics workflow for rodent biofluids, enabling a robust identification of metabolic alterations relevant to obesity and diabetes. The protocol bridges preclinical and clinical research by integrating standardised NMR acquisition, multivariate analysis, and rigorous statistical validation, which supports the discovery of key metabolic biomarkers and therapeutic targets.
5. Conclusions
These findings demonstrate the robustness and applicability of the described NMR-based metabolomics workflow for translational research, especially in preclinical rodent models of metabolic disease. The standardised methodology enables reproducible detection and quantification of clinically relevant metabolites, facilitating mechanistic insights and biomarker discovery. While optimised on the JEOL JNM-ECZ600R spectrometer, this workflow is adaptable to other major platforms such as Bruker, Agilent, and Varian through appropriate adjustments of instrument-specific parameters. Users should carefully consider hardware capabilities and operational differences, perform pilot calibrations, and thoroughly document acquisition settings to ensure reproducibility and cross-platform comparability. Looking ahead, adapting this workflow to additional sample matrices such as faecal material or incorporating stable isotope tracing techniques could provide a deeper understanding of complex biological processes, including gut–liver axis interactions and alterations in metabolic flux. It is important to note that this protocol does not include a comparative evaluation of different protein removal techniques or the utilisation of alternative experiments to assess their effects relative to the CPMG sequence. Such comparisons could offer valuable insights into optimising sample preparation and spectral acquisition tailored to specific research questions. Nevertheless, it is important to acknowledge the inherent limitations of NMR spectroscopy. Compared with mass spectrometry, NMR exhibits lower sensitivity and may encounter challenges due to spectral overlap in complex biological mixtures, limiting the detection of low-abundance and protein-bound metabolites. Therefore, the integration of NMR with complementary analytical platforms offers substantial potential for overcoming these constraints and achieving a more comprehensive, multidimensional metabolic profile. By combining the quantitative reliability and structural elucidation capabilities of NMR with the sensitivity and coverage of other techniques, future studies can leverage a synergistic approach that enhances metabolomic insights relevant to both preclinical and clinical settings.
Author Contributions
Conceptualization, M.N.M.N., T.K., M.F.M.N., Z.A.M.D. and S.S.N.; methodology, M.N.M.N., T.K., M.F.M.N., Z.A.M.D., S.S.N., E.F.L., M.F., S.Z.C.L. and A.Z.; formal analysis, M.N.M.N., T.K., M.F.M.N., Z.A.M.D., S.S.N. and K.C.; investigation, M.N.M.N., R.R., A.A., S.Z.C.L. and A.Z.; writing—original draft preparation, M.N.M.N., T.K., M.F.M.N., Z.A.M.D., S.S.N. and K.C.; writing—review and editing, R.R., E.F.L., M.F., A.A., S.Z.C.L. and A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Health, Malaysia (NMRR-20-413-5300).
Institutional Review Board Statement
This study was approved by the Animal Care and Usage Committee Ministry of Health, Malaysia (ACUC/KKM/02 (01/2021) and is registered with the Ministry of Health, Malaysia (NMRR-20-413-5300).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors upon reasonable request. A proposal with a detailed description of study objectives and a statistical analysis plan will be needed for an assessment of requests. Additional materials might also be required during the process of assessment.
Acknowledgments
The authors would like to thank the Director of the Institute for Medical Research (IMR) and the Director General of Health Malaysia for the permission to publish this paper. The authors would also like to thank all research team members from various institutions.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| 3D | Three-dimensional |
| ANOVA | Analysis of variance |
| CPMG | Carr–Purcell–Meiboom–Gill |
| DF | Degrees of freedom |
| FDR | False discovery rate |
| FID | Free induction decay |
| HMDB | Human Metabolome Database |
| HSD | Honest significant difference |
| KEGG | Kyoto Encyclopedia of Genomes and Genes |
| MVDA | Multivariate data analysis |
| NCD | Non-communicable disease |
| NMR | Nuclear magnetic resonance |
| PCA | Principal component analysis |
| PLS-DA | Partial least squares discriminant analysis |
| PRESAT | Presaturation |
| QC | Quality control |
| SNR | Signal-to-noise ratio |
| T2D | Type 2 diabetes |
| TSP | Sodium 3-(trimethylsilyl) propionic-2,2,3,3-d4 |
| VIP | Variable importance projection |
| ZDF | Zucker diabetic fatty |
References
- Nagana Gowda, G.A.; Raftery, D. NMR-Based Metabolomics. Adv. Exp. Med. Biol. 2021, 1280, 19–37. [Google Scholar] [CrossRef]
- Danzi, F.; Pacchiana, R.; Mafficini, A.; Scupoli, M.T.; Scarpa, A.; Donadelli, M.; Fiore, A. To metabolomics and beyond: A technological portfolio to investigate cancer metabolism. Signal Transduct. Target. Ther. 2023, 8, 137. [Google Scholar] [CrossRef]
- Debik, J.; Sangermani, M.; Wang, F.; Madssen, T.S.; Giskeødegård, G.F. Multivariate analysis of NMR-based metabolomic data. NMR Biomed. 2022, 35, e4638. [Google Scholar] [CrossRef]
- Stavarache, C.; Nicolescu, A.; Duduianu, C.; Ailiesei, G.L.; Balan-Porcăraşu, M.; Cristea, M.; Macsim, A.M.; Popa, O.; Stavarache, C.; Hîrtopeanu, A.; et al. A Real-Life Reproducibility Assessment for NMR Metabolomics. Diagnostics 2022, 12, 559. [Google Scholar] [CrossRef]
- Ndlovu, I.S.; Tshilwane, S.I.; Vosloo, A.; Chaisi, M.; Mukaratirwa, S. Metabolomics of Type 2 Diabetes Mellitus in Sprague Dawley Rats-In Search of Potential Metabolic Biomarkers. Int. J. Mol. Sci. 2023, 24, 12467. [Google Scholar] [CrossRef]
- Chowdhury, C.R.; Kavitake, D.; Jaiswal, K.K.; Jaiswal, K.S.; Reddy, G.B.; Agarwal, V.; Shetty, P.H. NMR-based metabolomics as a significant tool for human nutritional research and health applications. Food Biosci. 2023, 53, 102538. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Gholipourmalekabadi, M.; Shafikhani, S.H. Animal models for type 1 and type 2 diabetes: Advantages and limitations. Front. Endocrinol. 2024, 15, 1359685. [Google Scholar] [CrossRef]
- Shiota, M.; Printz, R.L. Diabetes in Zucker diabetic fatty rat. Methods Mol. Biol. 2012, 933, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.J.; Curtis, A.M.; Jen, A.; Thomson, J.A.; Clegg, D.O.; Jiang, P.; Coon, J.J.; Overmyer, K.A.; Toh, H. Plasma metabolomics supports non-fasted sampling for metabolic profiling across a spectrum of glucose tolerance in the Nile rat model for type 2 diabetes. Lab. Animal 2023, 52, 269–277. [Google Scholar] [CrossRef]
- Wishart, D.S.; Cheng, L.L.; Copié, V.; Edison, A.S.; Eghbalnia, H.R.; Hoch, J.C.; Gouveia, G.J.; Pathmasiri, W.; Powers, R.; Schock, T.B.; et al. NMR and Metabolomics-A Roadmap for the Future. Metabolites 2022, 12, 678. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Society. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ensor, J.E. Biomarker validation: Common data analysis concerns. Oncologist 2014, 19, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, T.A.H.; Von Rekowski, C.P.; Araújo, R.; Oliveira, M.C.; Justino, G.C.; Bento, L.; Calado, C.R.C. Comparison of two metabolomics-platforms to discover biomarkers in critically ill patients from serum analysis. Comput. Biol. Med. 2025, 184, 109393. [Google Scholar] [CrossRef]
- Ratitch, B.; Rodriguez-Chavez, I.R.; Dabral, A.; Fontanari, A.; Vega, J.; Onorati, F.; Vandendriessche, B.; Morton, S.; Damestani, Y. Considerations for Analyzing and Interpreting Data from Biometric Monitoring Technologies in Clinical Trials. Digit. Biomark. 2022, 6, 83–97. [Google Scholar] [CrossRef]
- Weinisch, P.; Fiamoncini, J.; Schranner, D.; Raffler, J.; Skurk, T.; Rist, M.J.; Römisch-Margl, W.; Prehn, C.; Adamski, J.; Hauner, H.; et al. Dynamic patterns of postprandial metabolic responses to three dietary challenges. Front. Nutr. 2022, 9, 933526. [Google Scholar] [CrossRef]
- Tien, D.S.Y.; Hockey, M.; So, D.; Stanford, J.; Clarke, E.D.; Collins, C.E.; Staudacher, H.M. Recommendations for Designing, Conducting, and Reporting Feeding Trials in Nutrition Research. Adv. Nutr. 2024, 15, 100283. [Google Scholar] [CrossRef]
- Brennan, L.; Hu, F.B.; Sun, Q. Metabolomics Meets Nutritional Epidemiology: Harnessing the Potential in Metabolomics Data. Metabolites 2021, 11, 709. [Google Scholar] [CrossRef] [PubMed]
- Menyhart, O.; Weltz, B.; Győrffy, B. MultipleTesting.com: A tool for life science researchers for multiple hypothesis testing correction. PLoS ONE 2021, 16, e0245824. [Google Scholar] [CrossRef]
- Otani, K.; Funada, H.; Teranishi, R.; Okada, M.; Yamawaki, H. Cardiovascular Characteristics of Zucker Fatty Diabetes Mellitus Rats, an Animal Model for Obesity and Type 2 Diabetes. Int. J. Mol. Sci. 2022, 23, 4228. [Google Scholar] [CrossRef]
- Yokoi, N.; Hoshino, M.; Hidaka, S.; Yoshida, E.; Beppu, M.; Hoshikawa, R.; Sudo, K.; Kawada, A.; Takagi, S.; Seino, S. A Novel Rat Model of Type 2 Diabetes: The Zucker Fatty Diabetes Mellitus ZFDM Rat. J. Diabetes Res. 2013, 2013, 103731. [Google Scholar] [CrossRef]
- Jeong, E.; Baek, Y.; Kim, H.-J.; Lee, H.G. Comparison of the anti-diabetic effects of various grain and legume extracts in high-fat diet and streptozotocin-nicotinamide-induced diabetic rats. Heliyon 2024, 10, e25279. [Google Scholar] [CrossRef]
- Arifin, W.N.; Zahiruddin, W.M. Sample Size Calculation in Animal Studies Using Resource Equation Approach. Malays. J. Med. Sci. 2017, 24, 101–105. [Google Scholar] [CrossRef]
- Ghini, V.; Abuja, P.M.; Polasek, O.; Kozera, L.; Laiho, P.; Anton, G.; Zins, M.; Klovins, J.; Metspalu, A.; Wichmann, H.E.; et al. Impact of the pre-examination phase on multicenter metabolomic studies. New Biotechnol. 2022, 68, 37–47. [Google Scholar] [CrossRef]
- Pauzi, F.A.; Sahathevan, S.; Khor, B.H.; Narayanan, S.S.; Zakaria, N.F.; Abas, F.; Karupaiah, T.; Daud, Z.A.M. Exploring Metabolic Signature of Protein Energy Wasting in Hemodialysis Patients. Metabolites 2020, 10, 291. [Google Scholar] [CrossRef] [PubMed]
- Madrid-Gambin, F.; Oller, S.; Marco, S.; Pozo, Ó.J.; Andres-Lacueva, C.; Llorach, R. Quantitative plasma profiling by 1H NMR-based metabolomics: Impact of sample treatment. Front. Mol. Biosci. 2023, 10, 1125582. [Google Scholar] [CrossRef] [PubMed]
- Kacerova, T.; Pires, E.; Walsby-Tickle, J.; Probert, F.; McCullagh, J.S.O. Integrating NMR and multi-LC-MS-based untargeted metabolomics for comprehensive analysis of blood serum samples. Anal. Chim. Acta 2025, 1356, 343979. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Raftery, D. Recent Advances in NMR-Based Metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.E.; Flott, T.L.; Schooff, C.R.; Smiley, Z.; Puskarich, M.A.; Myers, D.D.; Younger, J.G.; Jones, A.E.; Stringer, K.A. Rapid, Reproducible, Quantifiable NMR Metabolomics: Methanol and Methanol: Chloroform Precipitation for Removal of Macromolecules in Serum and Whole Blood. Metabolites 2018, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Barrilero, R.; Ramírez, N.; Vallvé, J.C.; Taverner, D.; Fuertes, R.; Amigó, N.; Correig, X. Unravelling and Quantifying the “NMR-Invisible” Metabolites Interacting with Human Serum Albumin by Binding Competition and T2 Relaxation-Based Decomposition Analysis. J. Proteome Res. 2017, 16, 1847–1856. [Google Scholar] [CrossRef]
- Barding, G.A., Jr.; Salditos, R.; Larive, C.K. Quantitative NMR for bioanalysis and metabolomics. Anal. Bioanal. Chem. 2012, 404, 1165–1179. [Google Scholar] [CrossRef]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed]
- Simón-Manso, Y.; Lowenthal, M.S.; Kilpatrick, L.E.; Sampson, M.L.; Telu, K.H.; Rudnick, P.A.; Mallard, W.G.; Bearden, D.W.; Schock, T.B.; Tchekhovskoi, D.V.; et al. Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal. Chem. 2013, 85, 11725–11731. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Quantitative NMR Methods in Metabolomics. In Handbook of Experimental Pharmacology; Springer International Publishing: Cham, Switzerland, 2023; Volume 277, pp. 143–164. [Google Scholar] [CrossRef]
- Saparuddin, F.; Mohd Nawi, M.N.; Ahmad Zamri, L.; Mansor, F.; Md Noh, M.F.; Omar, M.A.; Abdul Aziz, N.S.; Wahab, N.A.; Mediani, A.; Rajab, N.F.; et al. Metabolite, Biochemical, and Dietary Intake Alterations Associated with Lifestyle Interventions in Obese and Overweight Malaysian Women. Nutrients 2024, 16, 3501. [Google Scholar] [CrossRef]
- Ruhland, E.; Bund, C.; Outilaft, H.; Piotto, M.; Namer, I.J. A metabolic database for biomedical studies of biopsy specimens by high-resolution magic angle spinning nuclear MR: A qualitative and quantitative tool. Magn. Reson. Med. 2019, 82, 62–83. [Google Scholar] [CrossRef]
- Kim, J.A.; Choi, H.J.; Kwon, Y.K.; Ryu, D.H.; Kwon, T.H.; Hwang, G.S. 1H NMR-based metabolite profiling of plasma in a rat model of chronic kidney disease. PLoS ONE 2014, 9, e85445. [Google Scholar] [CrossRef]
- Xu, C.; Rezeng, C.; Li, J.; Zhang, L.; Yan, Y.; Gao, J.; Wang, Y.; Li, Z.; Chen, J. 1H NMR-Based Metabolomics Study of the Toxicological Effects in Rats Induced by “Renqing Mangjue” Pill, a Traditional Tibetan Medicine. Front. Pharmacol. 2017, 8, 602. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, Y.; Wu, D.; Wang, C.; Yang, Y.J.; Fan, S.J.; Xia, L.; Wei, Y.; Peng, X. (1)H-NMR metabolomics identifies significant changes in hypermetabolism after glutamine administration in burned rats. Am. J. Transl. Res. 2019, 11, 7286–7299. [Google Scholar]
- Jupin, M.; Michiels, P.J.; Girard, F.C.; Spraul, M.; Wijmenga, S.S. NMR metabolomics profiling of blood plasma mimics shows that medium- and long-chain fatty acids differently release metabolites from human serum albumin. J. Magn. Reson. 2014, 239, 34–43. [Google Scholar] [CrossRef]
- Laíns, I.; Duarte, D.; Barros, A.S.; Martins, A.S.; Gil, J.; Miller, J.B.; Marques, M.; Mesquita, T.; Kim, I.K.; Cachulo, M.d.L.; et al. Human plasma metabolomics in age-related macular degeneration (AMD) using nuclear magnetic resonance spectroscopy. PLoS ONE 2017, 12, e0177749. [Google Scholar] [CrossRef] [PubMed]
- Probert, F.; Ruiz-Rodado, V.; Vruchte, D.t.; Nicoli, E.-R.; Claridge, T.D.W.; Wassif, C.A.; Farhat, N.; Porter, F.D.; Platt, F.M.; Grootveld, M. NMR analysis reveals significant differences in the plasma metabolic profiles of Niemann Pick C1 patients, heterozygous carriers, and healthy controls. Sci. Rep. 2017, 7, 6320. [Google Scholar] [CrossRef] [PubMed]
- Hyberts, S.G.; Robson, S.A.; Wagner, G. Exploring signal-to-noise ratio and sensitivity in non-uniformly sampled multi-dimensional NMR spectra. J. Biomol. NMR 2013, 55, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Maldonado-Devincci, A.M.; Jiang, L. Evaluating line-broadening factors on a reference spectrum as a bucketing method for NMR based metabolomics. Anal. Biochem. 2020, 606, 113872. [Google Scholar] [CrossRef]
- Chenomx. Chenomx NMR Suite User Guide v9.02; Chenomx Inc.: Edmonton, AB, Canada, 2021. [Google Scholar]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).