An Optimized Protocol for SBEM-Based Ultrastructural Analysis of Cultured Human Cells
Abstract
1. Introduction
2. Materials and Methods
- Primary Dermal Fibroblast; Human, Neonatal - ATCC, Virginia, USA, Cat. No. PCS-201-010™;
- Essential 8™ Medium—Life Technologies, Waltham, MA, USA 02451, Cat No. A1517001;
- DPBS no calcium, no magnesium—Life Technologies, Waltham, MA, USA 02451, Cat No. 14190144;
- UltraPure™—0.5M EDTA, pH 8.0, Life Technologies, Waltham, MA, USA 02451, Cat No. 15575020.
- Glutaraldehyde—Sigma-Aldrich, Taufkirchen, Germany, Cat. No. G5882;
- Potassium hexacyanoferrate (III)—Warchem, Poland, Cat. No. 52731;
- Osmium tetroxide—Serva Cat. Heidelberg, Germany, No. 31251.03;
- Thiocarbohydrazide—Sigma-Aldrich, Taufkirchen, Germany, Cat. No. 223220;
- A 1% aqueous uranyl—SPI supplies, West Chester, USA Cat. No. 02624-AB;
- Lead nitrate—Supelco Taufkirchen, Germany, Cat. No. 1.07398;
- L- aspartic acid—Sigma-Aldrich, Taufkirchen, Germany, Cat. No. A9256;
- di-Sodium hydrogen phosphate dodecahydrate pure p.a.—Alchem, Poland, Cat. No. 363-117992809;
- Sodium dihydrogen phosphate monohydrate pure p.a.—Alchem, Poland, Cat. No. 363-117991804;
- Epoxy Embedding Medium—Polysciences, Hirschberg an der Bergstrasse, Germany Cat. No. 08792.
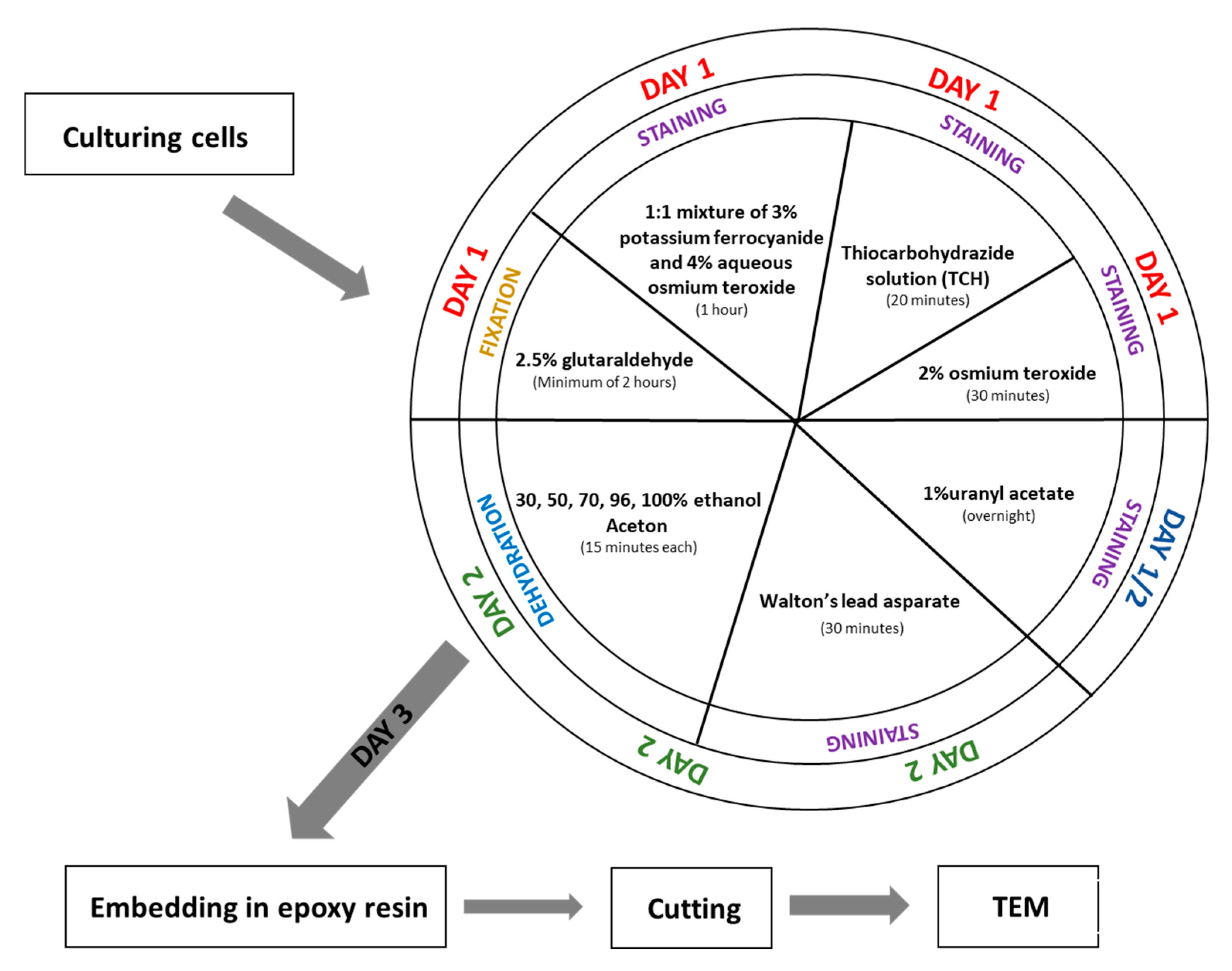
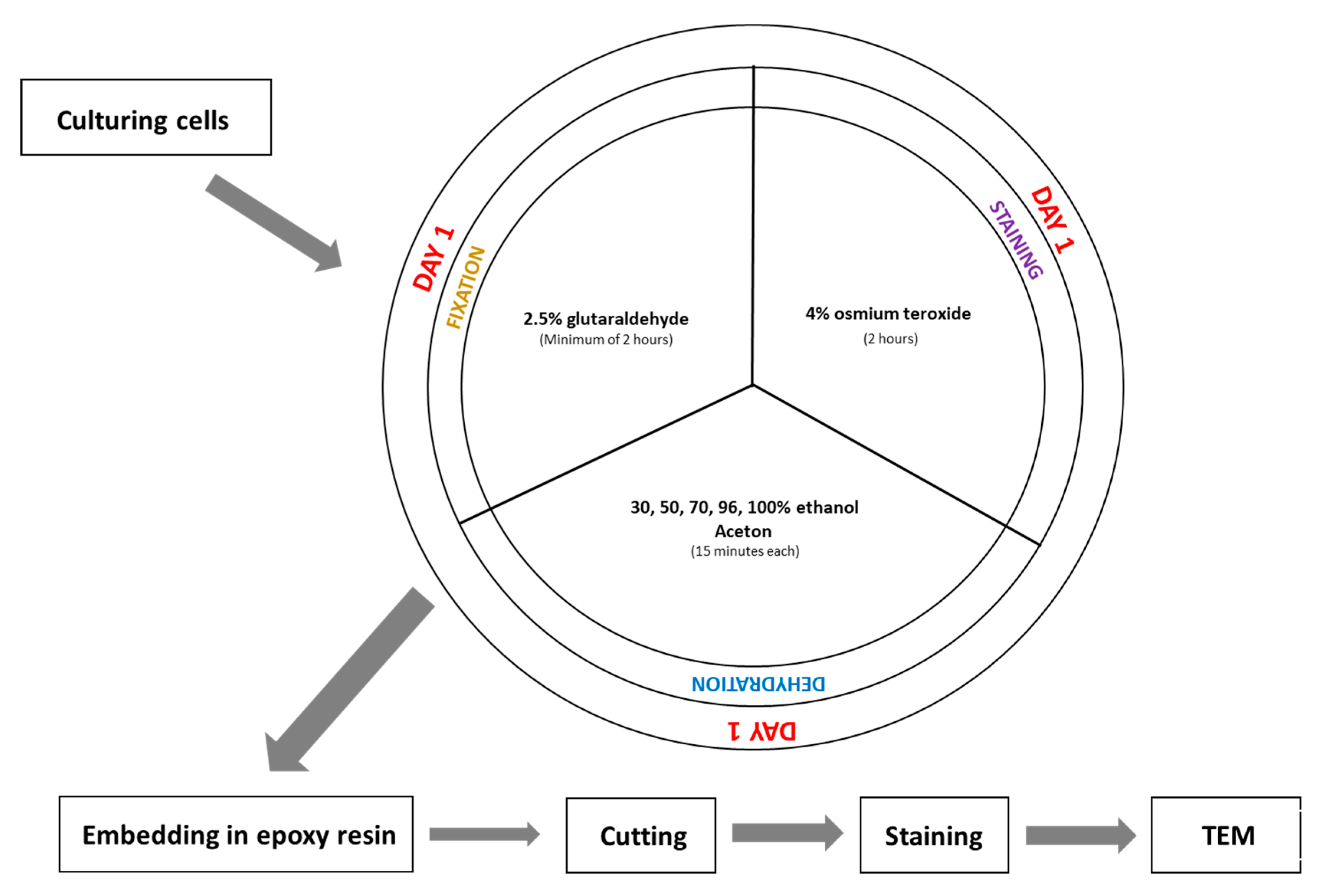
3. Procedure
- Fix the cell pellet in 2.5% glutaraldehyde prepared in 0.1 M phosphate buffer (pH 7.4) at room temperature for a minimum of 2 h. (The fixation time can be extended if necessary.)
- Wash the sample in 0.1 M phosphate buffer (pH 7.4) five times for 3 min each.(Note: If fixation time was extended, extend the washing steps accordingly.)
- Incubate the sample in a 1:1 mixture of 3% potassium ferrocyanide (in 0.1 M phosphate buffer) and 4% aqueous osmium tetroxide for 1 h on ice.
- While step 3 is ongoing, prepare the 1% thiocarbohydrazide (TCH) solution: Dissolve 0.1 g TCH in 10 mL of ddH2O. Incubate the solution at 60 °C for 1 h, swirling every 10 min to aid dissolution. Filter the solution through a 0.22 µm Millipore syringe filter. Note: TCH solution must always be freshly prepared before use.
- After step 3, wash the sample in ddH2O five times for 3 min each.
- Incubate the cell pellet in 1% TCH solution for 20 min at room temperature.
- Wash the sample again in ddH2O five times for 3 min each.
- Incubate the sample in 2% aqueous osmium tetroxide for 30 min at room temperature.
- Wash the sample in ddH2O five times for 3 min each.
- Add 1% aqueous uranyl acetate to the sample and incubate overnight at 4 °C (in the dark).
- Wash the sample in ddH2O five times for 3 min each at room temperature.
- While washing, prepare En bloc Walton’s lead aspartate solution. First, make a 0.03 M aspartic acid solution; this acid will dissolve faster as the pH will be lower. En bloc Walton’s lead aspartate solution consists of 0.066 g of lead nitrate in 10 mL of a 0.03 M aspartic acid solution. Adjust pH to 5.5 with 1 N KOH. Incubate the mixture at 60 °C for 30 min. While in incubation, no precipitate should form.
- Return to step 1. After incubation, wash the sample in ddH2O five times for 3 min each at room temperature.
- Add to the cells pellet En bloc Walton’s lead aspartate solution and incubate at 60 °C for 30 min.
- Wash the sample in ddH2O five times for 3 min each.
- Dehydrate the cell pellet in a graded ethanol series for 15 min at each step: 30%, 50%, 70%, 80%, and 96% ethanol, followed by four changes of 100% ethanol (15 min each).
- Incubate the sample for 15 min in a 1:1 solution of acetone and ethanol, then twice for 15 min in 100% acetone.
- After dehydration, transfer the samples to a 50% epoxy embedding medium in acetone and incubate for 2 h. Then, place the sample in an incubator at 56 °C overnight to allow the acetone to evaporate.
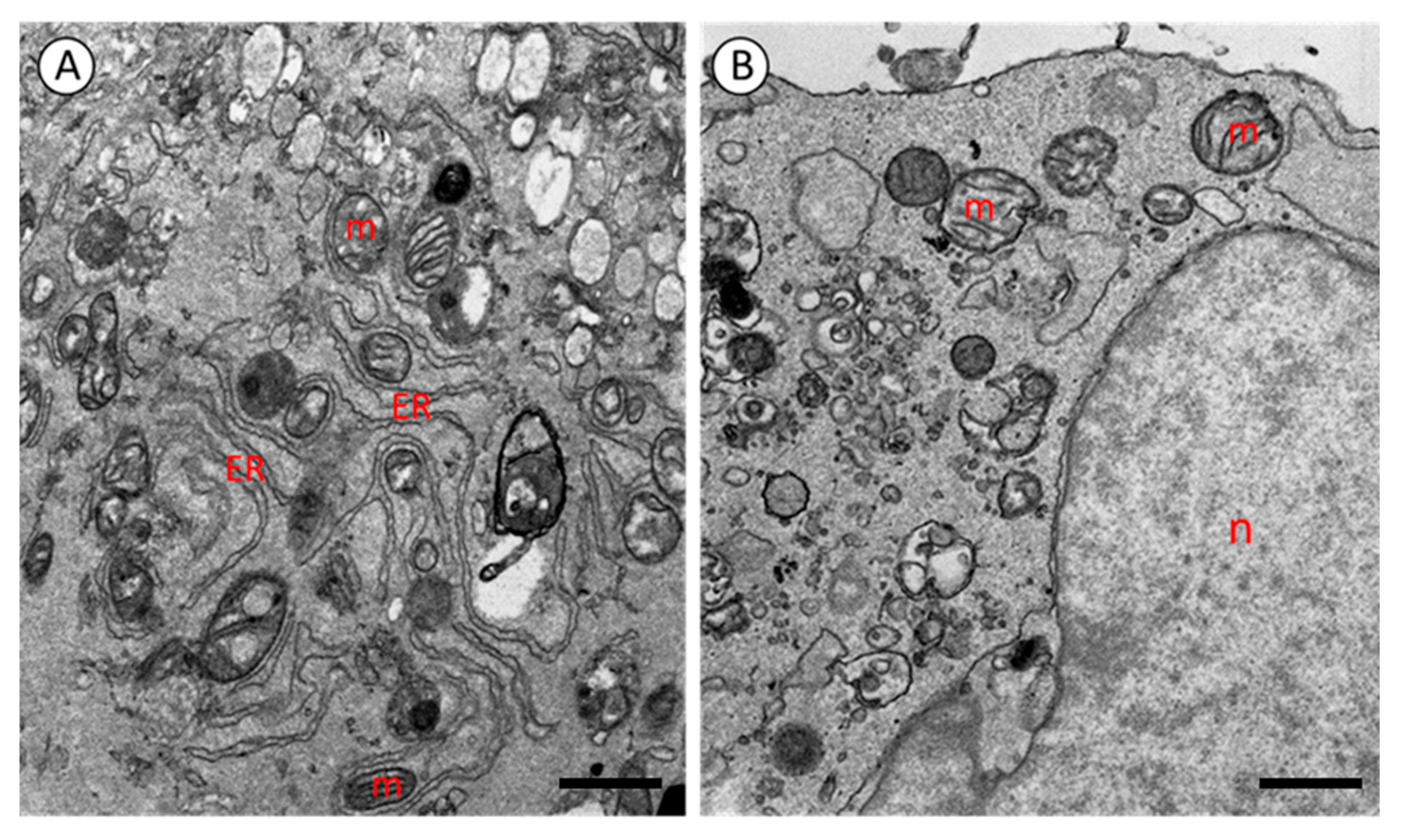
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CLEM | correlative light and electron microscopy |
| iPSCs | induced pluripotent stem cells |
| SBEM | serial block face electron microscopy |
| SEM | scanning electron microscopy |
| STEM | scanning transmission electron microscopy |
| TEM | transmission electron microscopy |
| TCH | thiocarbohydrazide |
References
- Elliott, A.D. Confocal microscopy: Principles and modern practices. Curr. Protoc. Cytom. 2020, 92, e68. [Google Scholar] [CrossRef] [PubMed]
- Micheva, K.D.; Smith, S.J. Array tomography: A new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron 2007, 200755, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Radulović, S.; Sunkara, S.; Rachel, R.; Leitinger, G. Three-dimensional SEM, TEM, and STEM for analysis of large-scale biological systems. Histochem. Cell Biol. 2022, 158, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Śliwińska, M.A.; Cały, A.; Szymański, J.; Radwańska, K. Serial Block-Face Scanning Electron Microscopy (SBEM) for the Study of Dendritic Spines. J. Vis. Exp. 2021, 176, e62712. [Google Scholar] [CrossRef] [PubMed]
- Antao, N.V.; Sall, J.; Petzold, C.; Ekiert, D.C.; Bhabha, G.; Liang, F.X. Sample preparationand data collection for serial block face scanning electron microscopy of mammalian cell monolayers. PLoS ONE 2024, 19, e0301284. [Google Scholar] [CrossRef] [PubMed]
- Antao, N.V.; Sall, J.; Petzold, C.; Ekiert, D.C.; Bhabha, G.; Liang, F.X. Sample preparation and data collection for serial block face scanning electron microscopy of mammalian cell monolayers. PLoS ONE 2024. [Google Scholar] [CrossRef] [PubMed]
- Liv, N.; Zonnevylle, A.C.; Narvaez, A.C.; Effting, A.P.; Voorneveld, P.W.; Lucas, M.S.; Hardwick, J.C.; Wepf, R.A.; Kruit, P.; Hoogenboom, J.P. Simultaneous correlative scanning electron and high-NA fluorescence microscopy. PLoS ONE 2013, 8, e55707. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.E.; Vasilescu, D.M.; Seal, K.A.D.; Keyes, S.D.; Mavrogordato, M.N.; Hogg, J.C.; Sinclair, I.; Warner, J.A.; Hackett, T.-L.; Lackie, P.M. Three Dimensional Imaging of Paraffin Embedded Human Lung Tissue Samples by Micro-Computed Tomography. PLoS ONE 2015, 10, e0126230. [Google Scholar] [CrossRef] [PubMed]
- Jansen, I.; Lucas, M.; Savci-Heijink, C.D.; Meijer, S.L.; Liem, E.I.M.L.; de Boer, O.J.; van Leeuwen, T.G.; Marquering, H.A.; de Bruin, D.M. Three-dimensional histopathological reconstruction of bladder tumours. Diagn. Pathol. 2019, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Onozato, M.L.; Klepeis, V.E.; Yagi, Y.; Mino-Kenudson, M. A role of three-dimensional (3D)-reconstruction in the classification of lung adenocarcinoma. Anal. Cell. Pathol. 2012, 35, 79–84. [Google Scholar] [CrossRef]
- Leighton, S.B. SEM images of block faces, cut by a miniature microtome within the SEM—A technical note. Scan Electron Microsc. 1981, Pt 2, 73–76. [Google Scholar]
- Denk, W.; Horstmann, H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004, 2, e329. [Google Scholar] [CrossRef] [PubMed]
- Hughes, L.; Hawes, C.; Monteith, S.; Vaughan, S. Serial block face scanning electron microscopy—The future of cell ultrastructure imaging. Protoplasma 2014, 251, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Deerinck, T.J.; Bushong, E.A.; Thor, A.K.; Ellisman, M.H. NCMIR methods for 3D EM: A new protocol for preparation of biological specimens for serial block face scanning electron microscopy. Microscopy [Online] 2010, 6. Available online: https://scispace.com/pdf/ncmir-methods-for-3d-em-a-new-protocol-for-preparation-of-3nyqnju13d.pdf (accessed on 25 July 2025).
- Diak, N.; Śliwińska, M.A.; Student, S.; Świątek, P. The three-dimensional conformation and activity of mitochondria in syncytial male germ line-cysts of medicinal leeches. Cell. Tissue Res. 2023, 394, 325–342. [Google Scholar] [CrossRef] [PubMed]
- Rost-Roszkowska, M.; Chajec, Ł.; Stojanović, D.; Antić, D. The ground pattern of midgut structure in Julidae (Julida: Juloidea): A study on selected species. Arthropod Syst. Phylogeny 2025, 83, 287–302. [Google Scholar] [CrossRef]
- Urbisz, A.Z.; Schmelz, R.M.; Małota, K.; Chajec, Ł.; Świątek, P. Conservative character of the germ-line cyst organization within enchytraeids (Annelida: Clitellata) ovary—New proofs based on two Achaeta species. Micron 2025, 188, 103732. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diak, N.; Chajec, Ł.; Fus-Kujawa, A.; Bajdak-Rusinek, K. An Optimized Protocol for SBEM-Based Ultrastructural Analysis of Cultured Human Cells. Methods Protoc. 2025, 8, 90. https://doi.org/10.3390/mps8040090
Diak N, Chajec Ł, Fus-Kujawa A, Bajdak-Rusinek K. An Optimized Protocol for SBEM-Based Ultrastructural Analysis of Cultured Human Cells. Methods and Protocols. 2025; 8(4):90. https://doi.org/10.3390/mps8040090
Chicago/Turabian StyleDiak, Natalia, Łukasz Chajec, Agnieszka Fus-Kujawa, and Karolina Bajdak-Rusinek. 2025. "An Optimized Protocol for SBEM-Based Ultrastructural Analysis of Cultured Human Cells" Methods and Protocols 8, no. 4: 90. https://doi.org/10.3390/mps8040090
APA StyleDiak, N., Chajec, Ł., Fus-Kujawa, A., & Bajdak-Rusinek, K. (2025). An Optimized Protocol for SBEM-Based Ultrastructural Analysis of Cultured Human Cells. Methods and Protocols, 8(4), 90. https://doi.org/10.3390/mps8040090






