Whole-Exome Sequencing Followed by dPCR-Based Personalized Genetic Approach in Solid Organ Transplantation: A Study Protocol and Preliminary Results
Abstract
1. Introduction
2. Materials and Methods
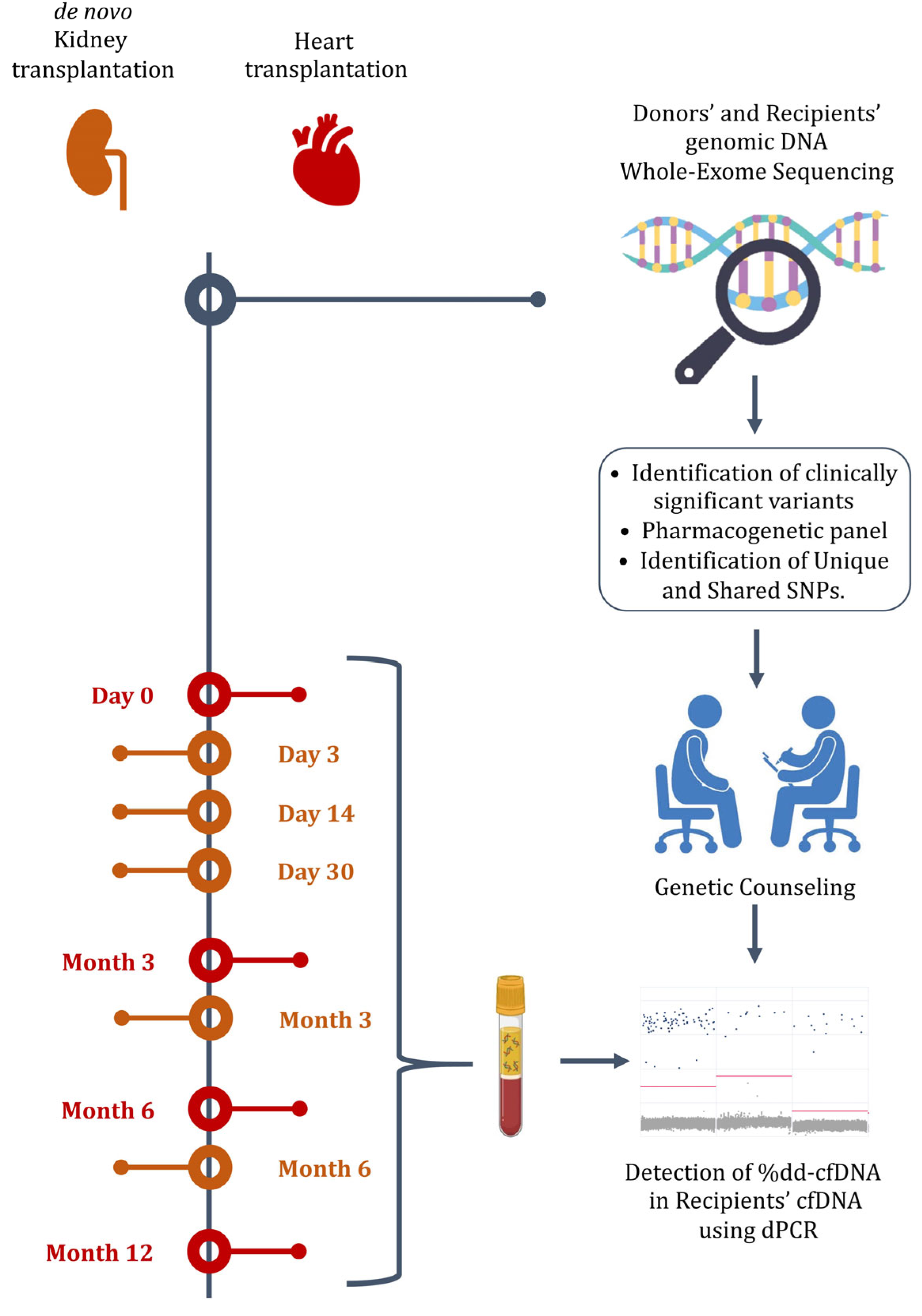
2.1. Study Design
2.2. Study Population and Eligibility Criteria
2.3. Participant Safety
2.4. Genomic and Cell-Free DNA Extraction
2.5. Whole-Exome Sequencing
2.6. Pharmacogenetic Analysis
2.7. Determination of dd-cfDNA%
2.8. Clinical Utility
2.9. Data Analysis
2.10. Ethical Considerations
3. Preliminary Results
3.1. Heart Transplantation
3.2. Adults Ans Pediatric Kidney Transplantation
3.3. Pharmacogenetics
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Nieuwenhuis, L.M.B.; Keating, B.J.; Festen, E.A.; de Meijer, V.E. The Impact of Donor and Recipient Genetic Variation on Outcomes After Solid Organ Transplantation: A Scoping Review and Future Perspectives. Transplantation 2022, 106, 1548–1557. [Google Scholar] [CrossRef]
- Roedder, S.; Vitalone, M.; Khatri, P.; Sarwal, M.M. Biomarkers in solid organ transplantation: Establishing personalized transplantation medicine. Genome Med. 2011, 3, 37. [Google Scholar] [CrossRef]
- Tantisattamo, E.; Reddy, U.G.; Ichii, H.; Ferrey, A.J.; Dafoe, D.C.; Ioannou, N.; Xie, J.; Pitman, T.R.; Hendricks, E.; Eguchi, N.; et al. Is It Time to Utilize Genetic Testing for Living Kidney Donor Evaluation? Nephron 2022, 146, 220–226. [Google Scholar] [CrossRef]
- Marin, E.P.; Cohen, E.; Dahl, N. Clinical Applications of Genetic Discoveries in Kidney Transplantation: A Review. Kidney360 2020, 1, 300–305. [Google Scholar] [CrossRef]
- Soraru, J.; Chakera, A.; Isbel, N.; Mallawaarachichi, A.; Rogers, N.; Trnka, P.; Patel, C.; Mallett, A.J. The Evolving Role of Diagnostic Genomics in Kidney Transplantation. Kidney Int. Rep. 2022, 7, 1758–1771. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, H.; Xiang, T.; Liu, D.; Xu, F.; Zhao, L.; Feng, Y.; Xu, L.; Liu, J.; Fang, Y.; et al. An accessible insight into genetic findings for transplantation recipients with suspected genetic kidney disease. NPJ Genom. Med. 2021, 6, 57. [Google Scholar] [CrossRef]
- Redondo, N.; Navarro, D.; Aguado, J.M.; Fernández-Ruiz, M. Human genetic polymorphisms and risk of viral infection after solid organ transplantation. Transplant. Rev. 2022, 36, 100669. [Google Scholar] [CrossRef]
- Boen, H.M.; Loeys, B.L.; Alaerts, M.; Saenen, J.B.; Goovaerts, I.; Van Laer, L.; Vorlat, A.; Vermeulen, T.; Franssen, C.; Pauwels, P.; et al. Diagnostic yield of genetic testing in heart transplant recipients with prior cardiomyopathy. J. Heart Lung Transplant. 2022, 41, 1218–1227. [Google Scholar] [CrossRef]
- Gourzi, P.; Pantou, M.P.; Gkouziouta, A.; Chaidaroglou, A.; Adamopoulos, S.; Degiannis, D. Can Genetic Profile of Patients Undergoing Heart Transplantation Alter Clinical Decisions? J. Heart Lung Transplant. 2020, 39, S232–S233. [Google Scholar] [CrossRef]
- Ettenger, R.; Albrecht, R.; Alloway, R.; Belen, O.; Cavaillé-Coll, M.W.; Chisholm-Burns, M.A.; Dew, M.A.; Fitzsimmons, W.E.; Nickerson, P.; Thompson, G.; et al. Meeting report: FDA public meeting on patient-focused drug development and medication adherence in solid organ transplant patients. Am. J. Transplant. 2018, 18, 564–573. [Google Scholar] [CrossRef]
- Deininger, K.M.; Tran, J.N.; Tsunoda, S.M.; Young, G.K.; Lee, Y.M.; Anderson, H.D.; Ii, R.L.P.; Hirsch, J.D.; Aquilante, C.L. Stakeholder perspectives of the clinical utility of pharmacogenomic testing in solid organ transplantation. Pharmacogenomics 2019, 20, 1291–1302. [Google Scholar] [CrossRef] [PubMed]
- Volpi, S.; Bult, C.J.; Chisholm, R.L.; Deverka, P.A.; Ginsburg, G.S.; Jacob, H.J.; Kasapi, M.; McLeod, H.L.; Roden, D.M.; Williams, M.S.; et al. Research Directions in the Clinical Implementation of Pharmacogenomics: An Overview of US Programs and Projects. Clin. Pharmacol. Ther. 2018, 103, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Rancic, N.; Dragojevic-Simic, V.; Vavic, N.; Kovacevic, A.; Segrt, Z.; Djordjevic, N. Economic Evaluation of Pharmacogenetic Tests in Patients Subjected to Renal Transplantation: A Review of Literature. Front. Public Health 2016, 4, 189. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Birdwell, K.A.; Decker, B.; Barbarino, J.M.; Peterson, J.F.; Stein, C.M.; Sadee, W.; Wang, D.; Vinks, A.A.; He, Y.; Swen, J.J.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin. Pharmacol. Ther. 2015, 98, 19–24. [Google Scholar] [CrossRef]
- PharmGKB. (n.d.). PharmGKB. Available online: https://www.pharmgkb.org/chemical/PA451578/prescribingInfo#fda-pgx-annotations (accessed on 25 October 2024).
- van Gelder, T.; van Schaik, R.H.; Hesselink, D.A. Pharmacogenetics and immunosuppressive drugs in solid organ transplantation. Nat. Rev. Nephrol. 2014, 10, 725–731. [Google Scholar] [CrossRef]
- Cascorbi, I. The Pharmacogenetics of Immune-Modulating Therapy. Adv. Pharmacol. 2018, 83, 275–296. [Google Scholar] [CrossRef]
- Thervet, E.; Anglicheau, D.; Legendre, C.; Beaune, P. Role of pharmacogenetics of immunosuppressive drugs in organ transplantation. Ther. Drug Monit. 2008, 30, 143–150. [Google Scholar] [CrossRef]
- Anglicheau, D.; Legendre, C.; Thervet, E. Pharmacogenetics in solid organ transplantation: Present knowledge and future perspectives. Transplantation 2004, 78, 311–315. [Google Scholar] [CrossRef]
- Elens, L.; Bouamar, R.; Shuker, N.; Hesselink, D.A.; van Gelder, T.; van Schaik, R.H. Clinical implementation of pharmacogenetics in kidney transplantation: Calcineurin inhibitors in the starting blocks. Br. J. Clin. Pharmacol. 2014, 77, 715–728. [Google Scholar] [CrossRef]
- Elens, L.; Hesselink, D.A.; van Schaik, R.H.; van Gelder, T. Pharmacogenetics in kidney transplantation: Recent updates and potential clinical applications. Mol. Diagn. Ther. 2012, 16, 331–345. [Google Scholar] [CrossRef]
- Brunet, M.; van Gelder, T.; Åsberg, A.; Haufroid, V.; Hesselink, D.A.; Langman, L.; Lemaitre, F.; Marquet, P.; Seger, C.; Shipkova, M.; et al. Therapeutic Drug Monitoring of Tacrolimus-Personalized Therapy: Second Consensus Report. Ther. Drug Monit. 2019, 41, 261–307. [Google Scholar] [CrossRef] [PubMed]
- Tron, C.; Lemaitre, F.; Verstuyft, C.; Petitcollin, A.; Verdier, M.C.; Bellissant, E. Pharmacogenetics of Membrane Transporters of Tacrolimus in Solid Organ Transplantation. Clin. Pharmacokinet. 2019, 58, 593–613. [Google Scholar] [CrossRef] [PubMed]
- Baimakhanov, B.B.; Chormanov, A.T.; Medeubekov, U.S.; Syrymov, Z.M.; Madadov, I.K.; Dabyltaeva, K.S.; Belgibaev, E.B.; Nabiev, E.S.; Saduakas, N.T.; Baiyz, A.Z.; et al. Genetic Polymorphism of Cyp3a5 as A Key Regulator of Pharmacokinetics of Tacrolimus in Kidney Transplant Patients: Evidence in Kazakh Population//Bulletin of Surgery in Kazakhstan. Вестник Хирургии Казахстана 2021, 1, 5–9. Available online: https://vhk.kz/wp-content/uploads/2021/05/1.pdf (accessed on 14 January 2024).
- Verhoeven, J.G.H.P.; Boer, K.; Van Schaik, R.H.N.; Manintveld, O.C.; Huibers, M.M.H.; Baan, C.C.; Hesselink, D.A. Liquid Biopsies to Monitor Solid Organ Transplant Function: A Review of New Biomarkers. Ther. Drug Monit. 2018, 40, 515–525. [Google Scholar] [CrossRef]
- Edwards, R.L.; Menteer, J.; Lestz, R.M.; Baxter-Lowe, L.A. Cell-free DNA as a solid-organ transplant biomarker: Technologies and approaches. Biomark. Med. 2022, 16, 401–415. [Google Scholar] [CrossRef]
- Preka, E.; Ellershaw, D.; Chandler, N.; Ahlfors, H.; Spencer, H.; Chitty, L.S.; Fenton, M.J.; Marks, S.D. Cell-Free DNA in Pediatric Solid Organ Transplantation Using a New Detection Method of Separating Donor-Derived from Recipient Cell-Free DNA. Clin. Chem. 2020, 66, 1300–1309. [Google Scholar] [CrossRef]
- Kataria, A.; Kumar, D.; Gupta, G. Donor-derived Cell-free DNA in Solid-organ Transplant Diagnostics: Indications, Limitations, and Future Directions. Transplantation 2021, 105, 1203–1211. [Google Scholar] [CrossRef]
- Bayanova, M.; Askerbekova, A.; Nazarova, L.; Abdikadirova, A.; Sapargaliyeva, M.; Malik, D.; Myrzakhmetova, G.; Pya Yu Bolatov, A. Genetic biomarkers of acute graft rejection after heart transplantation//Nauka i Zdravookhranenie. Sci. Healthc. 2024, 26, 177–189. [Google Scholar] [CrossRef]
- Filippone, E.J.; Farber, J.L. The Monitoring of Donor-derived Cell-free DNA in Kidney Transplantation. Transplantation 2021, 105, 509–516. [Google Scholar] [CrossRef]
- PharmGKB. Annotation of DPWG Guideline for Tacrolimus and CYP3A5. Available online: https://www.pharmgkb.org/chemical/PA451578/guidelineAnnotation/PA166104983 (accessed on 27 November 2024).
- PharmGKB. Annotation of RNPGx Guideline for Tacrolimus and CYP3A4, CYP3A5. Available online: https://www.pharmgkb.org/chemical/PA451578/guidelineAnnotation/PA166202481 (accessed on 27 November 2024).
- Bayanova, M.; Zhenissova, A.; Nazarova, L.; Abdikadirova, A.; Sapargalieyva, M.; Malik, D.; Bolatov, A.; Abdugafarov, S.; Assykbayev, M.; Altynova, S.; et al. Influence of Genetic Polymorphisms in CYP3A5, CYP3A4, and MDR1 on Tacrolimus Metabolism after kidney transplantation. J. Clin. Med. Kazakhstan 2024, 21, 11–17. [Google Scholar] [CrossRef]
- Brunet, M.; Pastor-Anglada, M. Insights into the Pharmacogenetics of Tacrolimus Pharmacokinetics and Pharmacodynamics. Pharmaceutics 2022, 14, 1755. [Google Scholar] [CrossRef] [PubMed]
- Benning, L.; Morath, C.; Fink, A.; Rudek, M.; Speer, C.; Kälble, F.; Nusshag, C.; Beimler, J.; Schwab, C.; Waldherr, R.; et al. Donor-Derived Cell-Free DNA (dd-cfDNA) in Kidney Transplant Recipients with Indication Biopsy-Results of a Prospective Single-Center Trial. Transpl. Int. 2023, 36, 11899. [Google Scholar] [CrossRef] [PubMed]
- Dandamudi, R.; Gu, H.; Goss, C.W.; Walther, L.; Dharnidharka, V.R. Longitudinal Evaluation of Donor-Derived Cellfree DNA in Pediatric Kidney Transplantation. Clin. J. Am. Soc. Nephrol. 2022, 17, 1646–1655. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, Q.; Cui, Z.; Wang, Z.; Zeng, F.; Zhang, Y. Tacrolimus Concentration Is Effectively Predicted Using Combined Clinical and Genetic Factors in the Perioperative Period of Kidney Transplantation and Associated with Acute Rejection. J. Immunol. Res. 2022, 2022, 3129389. [Google Scholar] [CrossRef]
- Azam, F.; Khan, M.; Khaliq, T.; Bhatti, A.H. Influence of ABCB1 gene polymorphism on concentration to dose ratio and adverse effects of tacrolimus in Pakistani liver transplant recipients. Pak. J. Med. Sci. 2021, 37. [Google Scholar] [CrossRef]
- Dessilly, G.; Elens, L.; Panin, N.; Capron, A.; Decottignies, A.; Demoulin, J.B.; Haufroid, V. ABCB1 1199G>A genetic polymorphism (Rs2229109) influences the intracellular accumulation of tacrolimus in HEK293 and K562 recombinant cell lines. PLoS ONE 2014, 9, e91555. [Google Scholar] [CrossRef]
- Huang, S.; Song, W.; Jiang, S.; Li, Y.; Wang, M.; Yang, N.; Zhu, H. Pharmacokinetic interactions between tacrolimus and Wuzhi capsule in liver transplant recipients: Genetic polymorphisms affect the drug interaction. Chem.-Biol. Interact. 2024, 391, 110906. [Google Scholar] [CrossRef]
- Genvigir, F.D.V.; Nishikawa, A.M.; Felipe, C.R.; Tedesco-Silva, H., Jr.; Oliveira, N.; Salazar, A.B.C.; Medina-Pestana, J.O.; Doi, S.Q.; Hirata, M.H.; Hirata, R.D.C. Influence of ABCC2, CYP2C8, and CYP2J2 Polymorphisms on Tacrolimus and Mycophenolate Sodium-Based Treatment in Brazilian Kidney Transplant Recipients. Pharmacotherapy 2017, 37, 535–545. [Google Scholar] [CrossRef]
- Dong, Y.; Xu, Q.; Li, R.; Tao, Y.; Zhang, Q.; Li, J.; Ma, Z.; Shen, C.; Zhong, M.; Wang, Z.; et al. CYP3A7, CYP3A4, and CYP3A5 genetic polymorphisms in recipients rather than donors influence tacrolimus concentrations in the early stages after liver transplantation. Gene 2022, 809, 146007. [Google Scholar] [CrossRef]
- Tavira, B.; Coto, E.; Torres, A.; Díaz-Corte, C.; Díaz-Molina, B.; Ortega, F.; Arias, M.; Díaz, J.M.; Selgas, R.; López-Larrea, C.; et al. Pharmacogenetics of tacrolimus REDINREN study group Association between a common KCNJ11 polymorphism (rs5219) and new-onset posttransplant diabetes in patients treated with Tacrolimus. Mol. Genet. Metab. 2012, 105, 525–527. [Google Scholar] [CrossRef]
- Kurzawski, M.; Malinowski, D.; Dziewanowski, K.; Droździk, M. Analysis of common polymorphisms within NR1I2 and NR1I3 genes and tacrolimus dose-adjusted concentration in stable kidney transplant recipients. Pharmacogenet. Genom. 2017, 27, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, R.X.; Li, J.; Zhang, Y.; Wang, X.D.; Fu, Q.; Chen, L.Y.; Liu, X.M.; Huang, H.B.; Huang, M.; et al. The POR rs1057868-rs2868177 GC-GT diplotype is associated with high tacrolimus concentrations in early post-renal transplant recipients. Acta Pharmacol. Sin. 2016, 37, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, Y.; Hu, Y.; Zhang, X.; Zhong, L.; Fan, J.; Peng, Z. Association of donor and recipient SUMO4 rs237025 genetic variant with new-onset diabetes mellitus after liver transplantation in a Chinese population. Gene 2017, 627, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.H.; Zhang, Q. Clinical applications of next-generation sequencing in histocompatibility and transplantation. Curr. Opin. Organ Transplant. 2015, 20, 461–467. [Google Scholar] [CrossRef]
- Liu, C.; Yang, X. Using Exome and Amplicon-Based Sequencing Data for High-Resolution HLA Typing with ATHLATES. Methods Mol. Biol. 2018, 1802, 203–213. [Google Scholar] [CrossRef]
- Petersdorf, E.W.; Stevenson, P.; Malkki, M.; Strong, R.K.; Spellman, S.R.; Haagenson, M.D.; Horowitz, M.M.; Gooley, T.; Wang, T. Patient HLA Germline Variation and Transplant Survivorship. J. Clin. Oncol. 2018, 36, 2524–2531. [Google Scholar] [CrossRef]
- Cornaby, C.; Schmitz, J.L.; Weimer, E.T. Next-generation sequencing and clinical histocompatibility testing. Hum. Immunol. 2021, 82, 829–837. [Google Scholar] [CrossRef]
- Koelemen, J.; Gotthardt, M.; Steinmetz, L.M.; Meder, B. RBM20-Related Cardiomyopathy: Current Understanding and Future Options. J. Clin. Med. 2021, 10, 4101. [Google Scholar] [CrossRef]
- Novelli, V.; Malkani, K.; Cerrone, M. Pleiotropic Phenotypes Associated with PKP2 Variants. Front. Cardiovasc. Med. 2018, 5, 184. [Google Scholar] [CrossRef]
- Brown, E.E.; Murray, B.; Vaishnav, J.; Tampakakis, E.; Barouch, L.A.; James, C.; Murphy, A.M.; Judge, D.P. Genetic Dilated Cardiomyopathy Due to TTN Variants Without Known Familial Disease. Circulation. Genom. Precis. Med. 2020, 13, e003082. [Google Scholar] [CrossRef]
- Clausen, F.B.; Jørgensen, K.M.C.L.; Wardil, L.W.; Nielsen, L.K.; Krog, G.R. Droplet digital PCR-based testing for donor-derived cell-free DNA in transplanted patients as noninvasive marker of allograft health: Methodological aspects. PLoS ONE 2023, 18, e0282332. [Google Scholar] [CrossRef] [PubMed]
- Jerič Kokelj, B.; Štalekar, M.; Vencken, S.; Dobnik, D.; Kogovšek, P.; Stanonik, M.; Arnol, M.; Ravnikar, M. Feasibility of Droplet Digital PCR Analysis of Plasma Cell-Free DNA From Kidney Transplant Patients. Front. Med. 2021, 8, 748668. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, L.; Westerling, S.; Talla, V.; Sendel, A.; Wennberg, L.; Olsson, R.; Hedrum, A.; Hauzenberger, D. Development and performance of a next generation sequencing (NGS) assay for monitoring of dd-cfDNA post solid organ transplantation. Clin. Chim. Acta Int. J. Clin. Chem. 2024, 552, 117647. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.S.; Almokayad, I.; Collins, A.; Raj, D.; Jagadeesan, M. Donor-derived Cell-free DNA: Advancing a Novel Assay to New Heights in Renal Transplantation. Transplant. Direct 2021, 7, e664. [Google Scholar] [CrossRef]
- Semenova, Y.; Bayanova, M.; Rakhimzhanova, S.; Altynova, S.; Sailybayeva, A.; Asanova, A.; Pya, Y. Understanding Pediatric Kidney Transplant Rejection: Its Pathophysiology, Biomarkers, and Management Strategies. Curr. Med. Chem. 2024, 31, 1–20. [Google Scholar] [CrossRef]
- Agbor-Enoh, S.; Shah, P.; Tunc, I.; Hsu, S.; Russell, S.; Feller, E.; Shah, K.; Rodrigo, M.E.; Najjar, S.S.; Kong, H.; et al. GRAfT Investigators Cell-Free DNA to Detect Heart Allograft Acute Rejection. Circulation 2021, 143, 1184–1197. [Google Scholar] [CrossRef]
- Khush, K.K.; Patel, J.; Pinney, S.; Kao, A.; Alharethi, R.; DePasquale, E.; Ewald, G.; Berman, P.; Kanwar, M.; Hiller, D.; et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: A prospective multicenter study. Am. J. Transplant. 2019, 19, 2889–2899. [Google Scholar] [CrossRef]
- Böhmer, J.; Wasslavik, C.; Andersson, D.; Ståhlberg, A.; Jonsson, M.; Wåhlander, H.; Karason, K.; Sunnegårdh, J.; Nilsson, S.; Asp, J.; et al. Absolute Quantification of Donor-Derived Cell-Free DNA in Pediatric and Adult Patients After Heart Transplantation: A Prospective Study. Transpl. Int. 2023, 36, 11260. [Google Scholar] [CrossRef]
- Cucchiari, D.; Cuadrado-Payan, E.; Gonzalez-Roca, E.; Revuelta, I.; Argudo, M.; Ramirez-Bajo, M.J.; Ventura-Aguiar, P.; Rovira, J.; Bañon-Maneus, E.; Montagud-Marrahi, E.; et al. Early kinetics of donor-derived cell-free DNA after transplantation predicts renal graft recovery and long-term function. Nephrol. Dial. Transplant. 2023, 39, 114–121. [Google Scholar] [CrossRef]
- De Vlaminck, I.; Martin, L.; Kertesz, M.; Patel, K.; Kowarsky, M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl. Acad. Sci. USA 2015, 112, 13336–13341. [Google Scholar] [CrossRef]
- Knight, S.R.; Thorne, A.; Lo Faro, M.L. Donor-specific Cell-free DNA as a Biomarker in Solid Organ Transplantation. A Systematic Review. Transplantation 2019, 103, 273–283. [Google Scholar] [CrossRef]
| Case | Diagnosis | Age at HTx (Year) | Time (Month) | Genetic Variant | ACMG | dd-cfDNA % | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | M ± SD | F, p | ||||||
| HTx1 | DCM | 33 | 79 | TTN:c.29338A > G (p.Ile9780Val) | VUS | 0.04% | 0.04% | 0.01% | 0.0001% | 0.023 ± 0.021 | 34.0, p = 0.003 Post-hoc test: HTx2 vs. HTx1/HTx3, p < 0.05 |
| HTx2 | DCM | 26 | 104 | RBM20:c.1907G > A (p.Arg636His) | P | 0.70% | 0.60% | 0.80% | 0.46% | 0.640 ± 0.145 | |
| HTx3 | HCM | 46 | 25 | PKP2:c.288T > G (p.Asp96Glu) | VUS | 0.008% | 0.006% | 0.002% | 0.005 ± 0.003 | ||
| Case | Diagnosis | Age at KTx (Year) | Donor Type | dd-cfDNA % | ||||
|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | T5 | ||||
| AKTx1 | Glomerular disease | 66 | LDKT | 0.17% | 0.08% | 0.13% | 0.03% | 0.002% |
| AKTx2 | CKD | 51 | DDKT | 12.0% | 0.60% | 0.20% | - | 0.10% |
| AKTx3 | Glomerular disease | 57 | LDKT | 0.24% | 0.17% | 0.06% | 0.09% | 0.25% |
| AKTx4 | Glomerular disease | 43 | LDKT | 0.17% | 0.45% | 0.12% | - | - |
| AKTx5 | Glomerular disease | 33 | LDKT | 0.30% | 0.02% | - | - | - |
| PKTx1 | CAKUT | 17 | LDKT | 0.07% | 0.17% | 0.19% | 0.23% | 0.23% |
| PKTx2 | CAKUT | 7 | LDKT | 1.72% | 7.35% | - | 0.51% | - |
| Genetic Variant (rs) | Gene | Distribution Among 10 Study Participants | Effect | Reference |
|---|---|---|---|---|
| rs1045642 | ABCB1 | 6 heterozygous and 2 homozygous cases | SNPs were found to have a potential effect on early tacrolimus C0/D | [37] |
| rs2032582 | ABCB1 | 5 heterozygous and 3 homozygous cases | ||
| rs1128503 | ABCB1 | 4 heterozygous and 3 homozygous cases | Association with acute cellular rejection | [38] |
| rs2229109 | ABCB1 | 1 heterozygous case | Association between SNP and tacrolimus intracellular accumulation | [39] |
| rs3740066 | ABCC2 | 3 heterozygous cases | SNP was found to have potential effect on early tacrolimus C 0/D | [37] |
| rs717620 | ABCC2 | 1 heterozygous case | Significant factor of tacrolimus lnC/D among LTx recipients | [40] |
| rs890293 | CYP2J2 | 1 heterozygous case | Influenced the renal function of these patients and the occurrence of adverse events during treatment with tacrolimus among KTx recipients | [41] |
| rs2242480 | CYP3A4 | 1 heterozygous case | Carriers had an almost twofold increase in the tacrolimus C0/D compared to that of the non-carriers | [42] |
| rs5219 | KCNJ11 | 4 heterozygous cases | Polymorphism associated with a new-onset posttransplant diabetes in patients treated with tacrolimus | [43] |
| rs2276707 | NR1I2 | 5 heterozygous cases | Impact tacrolimus clearance in kidney and liver transplant recipients | [44] |
| rs1057868 | POR | 4 heterozygous and 1 homozygous cases | SNPs rs1057868-rs2868177 GC-GT diplotype is associated with high tacrolimus concentrations in early post-renal transplant recipients | [45] |
| rs237025 | SUMO4 | 8 heterozygous and 1 homozygous cases | SNP contributes to the development of new-onset diabetes mellitus after liver transplantation | [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayanova, M.; Bolatov, A.; Malik, D.; Zhenissova, A.; Abdikadirova, A.; Sapargaliyeva, M.; Nazarova, L.; Myrzakhmetova, G.; Novikova, S.; Turganbekova, A.; et al. Whole-Exome Sequencing Followed by dPCR-Based Personalized Genetic Approach in Solid Organ Transplantation: A Study Protocol and Preliminary Results. Methods Protoc. 2025, 8, 27. https://doi.org/10.3390/mps8020027
Bayanova M, Bolatov A, Malik D, Zhenissova A, Abdikadirova A, Sapargaliyeva M, Nazarova L, Myrzakhmetova G, Novikova S, Turganbekova A, et al. Whole-Exome Sequencing Followed by dPCR-Based Personalized Genetic Approach in Solid Organ Transplantation: A Study Protocol and Preliminary Results. Methods and Protocols. 2025; 8(2):27. https://doi.org/10.3390/mps8020027
Chicago/Turabian StyleBayanova, Mirgul, Aidos Bolatov, Dias Malik, Aida Zhenissova, Aizhan Abdikadirova, Malika Sapargaliyeva, Lyazzat Nazarova, Gulzhan Myrzakhmetova, Svetlana Novikova, Aida Turganbekova, and et al. 2025. "Whole-Exome Sequencing Followed by dPCR-Based Personalized Genetic Approach in Solid Organ Transplantation: A Study Protocol and Preliminary Results" Methods and Protocols 8, no. 2: 27. https://doi.org/10.3390/mps8020027
APA StyleBayanova, M., Bolatov, A., Malik, D., Zhenissova, A., Abdikadirova, A., Sapargaliyeva, M., Nazarova, L., Myrzakhmetova, G., Novikova, S., Turganbekova, A., & Pya, Y. (2025). Whole-Exome Sequencing Followed by dPCR-Based Personalized Genetic Approach in Solid Organ Transplantation: A Study Protocol and Preliminary Results. Methods and Protocols, 8(2), 27. https://doi.org/10.3390/mps8020027






