Radiogenomics, Breast Cancer Diagnosis and Characterization: Current Status and Future Directions
Abstract
:1. Introduction
2. Biomarkers for BC
2.1. Biochemical Markers: Advantages and Limitations
- Circulating tumor DNA (ctDNA) or circulating free DNA (cfDNA) are small fragments released in the blood system from the primary tumor or metastatic cells. They are DNA fragments less than 500 bp in length and exhibit the same somatic alteration present in the tumor from which they originate, including point mutations, chromosomal rearrangements, copy number variations, and DNA methylations. As the amount of cfDNA is very small, the detection method used for their quantitation is mainly based on polymerase chain reaction (PCR) amplification and next-generation sequencing (NGS). These DNA fragments could be actively released via microvesicle release or the degradation of apoptotic and necrotic cancer cells [22]. From the original discovery of cfDNA in 1948, several papers have demonstrated that these DNA fragments originated from tumor cells undergoing genomic instability. In 1994, it was demonstrated that these small DNA showed the same specific genomic mutation of the primary tumor [23,24]. cfDNA obtained by plasma isolation at different time points could be helpful in the description of the natural course of cancer development before and after therapeutic treatment.
- Circulating tumor cells (CTCs). The presence of disseminating tumor cells is a common feature of solid cancer, such as BC. The detection of these cells is associated with poor outcomes at the level of both overall survival and disease-free survival in BC [25]. Disseminated tumor cells are usually isolated from a patient’s bone marrow, with an invasive technique that is not always accepted by the patients. CTCs are epithelial cells released by the primary tumor in the number of less than 100 cells per ml of peripheral blood. They are able to differentiate cancer patients from healthy subjects [26].
- Non-coding RNAs (ncRNA). ncRNAs are crucial regulators of gene expression and are strongly associated with BC. The large family of ncRNAs includes several regulatory RNAs, such as microRNA (miRNAs), long non-coding RNAs (lncRNA), and circular RNAs (circRNAs). miRNAs are small RNAs of 19–25 nucleotides able to regulates the mRNA profiles inside each cells; they could also be secreted in the microenvironment of the tumor as well as in body biofluids (blood, lacrimae, urine, etc.). The list of miRNAs have been deposited into miRBse database (https://www.mirbase.org, accessed on 1 July 2022) [27], which annotated more than 38,500 predicted miRNA sequences (release V22.1).
2.2. BC imaging Biomarkers: From Standard Quantification to Radiomics
3. Radiogenomics: Combining Molecular and Imaging Biomarkers for BC Characterization
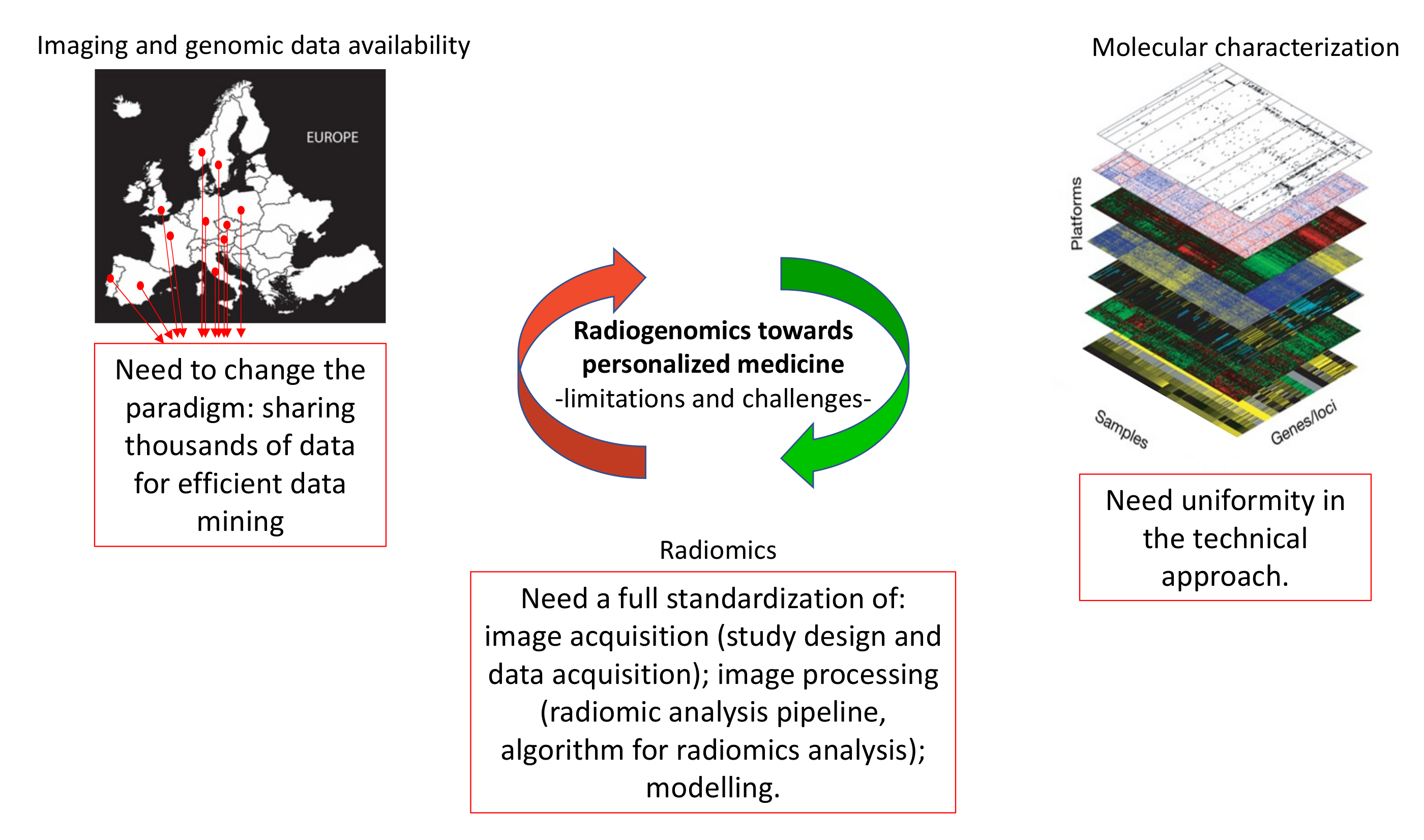
4. Limitations of Radiogenomic Approach
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Tilli, T.M. Precision Medicine: Technological Impact into Breast Cancer Diagnosis, Treatment and Decision Making. J. Pers. Med. 2021, 11, 1348. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Jiang, N.; Wu, Y. Application and Analysis of Biomedical Imaging Technology in Early Diagnosis of Breast Cancer. Methods Mol. Biol. 2020, 2204, 63–73. [Google Scholar] [PubMed]
- Jiang, M.; Lei, S.; Zhang, J.; Hou, L.; Zhang, M.; Luo, Y. Multimodal Imaging of Target Detection Algorithm under Artificial Intelligence in the Diagnosis of Early Breast Cancer. J. Health. Eng. 2022, 2022, 9322937. [Google Scholar] [CrossRef]
- Paydary, K.; Seraj, S.M.; Zadeh, M.Z.; Emamzadehfard, S.; Shamchi, S.P.; Gholami, S.; Werner, T.J.; Alavi, A. The Evolving Role of FDG-PET/CT in the Diagnosis, Staging, and Treatment of Breast Cancer. Mol. Imaging Biol. 2019, 21, 1–10. [Google Scholar] [CrossRef]
- Guo, R.; Lu, G.; Qin, B.; Fei, B. Ultrasound Imaging Technologies for Breast Cancer Detection and Management: A Review. Ultrasound Med. Biol. 2018, 44, 37–70. [Google Scholar] [CrossRef]
- Sarikaya, I. Breast Cancer and PET Imaging. Nucl. Med. Rev. Cent. East Eur. 2021, 24, 16–26. [Google Scholar] [CrossRef]
- Leithner, D.; Wengert, G.J.; Helbich, T.H.; Thakur, S.; Ochoa-Albiztegui, R.E.; Morris, E.A.; Pinker, K. Clinical role of breast MRI now and going forward. Clin. Radiol. 2018, 73, 700–714. [Google Scholar] [CrossRef]
- Tabouret-Viaud, C.; Botsikas, D.; Delattre, B.M.; Mainta, I.; Amzalag, G.; Rager, O.; Vinh-Hung, V.; Miralbell, R.; Ratib, O. PET/MR in Breast Cancer. Semin. Nucl. Med. 2015, 45, 304–321. [Google Scholar] [CrossRef]
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020, 471, 61–71. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Segal, E.; Sirlin, C.B.; Ooi, C.; Adler, A.S.; Gollub, J.; Chen, X.; Chan, B.K.; Matcuk, G.R.; Barry, C.T.; Chang, H.Y.; et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat. Biotechnol. 2007, 25, 675–680. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [Green Version]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Shiradkar, R.; Liu, Z. Integrating pathomics with radiomics and genomics for cancer prognosis: A brief review. Chin. J. Cancer Res. 2021, 33, 563–573. [Google Scholar] [CrossRef]
- Zhang, Q.; Fu, Q.; Bai, X.; Liang, T. Molecular Profiling-Based Precision Medicine in Cancer: A Review of Current Evidence and Challenges. Front. Oncol. 2020, 10, 532403. [Google Scholar] [CrossRef]
- Yu, K.H.; Snyder, M. Omics Profiling in Precision Oncology. Mol. Cell Proteomics 2016, 15, 2525–2536. [Google Scholar] [CrossRef] [Green Version]
- Eliyatkin, N.; Yalcin, E.; Zengel, B.; Aktas, S.; Vardar, E. Molecular Classification of Breast Carcinoma: From Traditional, Old-Fashioned Way to A New Age, and A New Way. J. Breast Health 2015, 11, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Sun, N.; Gao, P.; Li, Y.; Yan, Z.; Peng, Z.; Zhang, Y.; Han, F.; Qi, X. Screening and Identification of Key Common and Specific Genes and Their Prognostic Roles in Different Molecular Subtypes of Breast Cancer. Front. Mol. Biosci. 2021, 8, 619110. [Google Scholar] [CrossRef]
- Burstein, H.J.; Curigliano, G.; Thurlimann, B.; Weber, W.P.; Poortmans, P.; Regan, M.M.; Senn, H.J.; Winer, E.P.; Gnant, M.; Panelists of the St Gallen Consensus Conference. Customizing local and systemic therapies for women with early breast cancer: The St. Gallen International Consensus Guidelines for treatment of early breast cancer 2021. Ann. Oncol. 2021, 32, 1216–1235. [Google Scholar] [CrossRef]
- Fernandez-Lazaro, D.; Garcia Hernandez, J.L.; Garcia, A.C.; Cordova Martinez, A.; Mielgo-Ayuso, J.; Cruz-Hernandez, J.J. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, K.C.S.; Ramos, I.B.; Silva, J.M.C.; Barra, W.F.; Riggins, G.J.; Palande, V.; Pinho, C.T.; Frenkel-Morgenstern, M.; Santos, S.E.B.; Assumpcao, P.P.; et al. Current Perspectives on Circulating Tumor DNA, Precision Medicine, and Personalized Clinical Management of Cancer. Mol. Cancer Res. 2020, 18, 517–528. [Google Scholar] [CrossRef] [Green Version]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomark. Prev. 1994, 3, 67–71. [Google Scholar]
- Vasioukhin, V.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Stroun, M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br. J. Haematol. 1994, 86, 774–779. [Google Scholar] [CrossRef]
- Hartkopf, A.D.; Banys, M.; Krawczyk, N.; Wallwiener, M.; Schneck, H.; Neubauer, H.; Fehm, T. Circulating Tumor Cells in Early-Stage Breast Cancer. Geburtshilfe Frauenheilkd 2011, 71, 1067–1072. [Google Scholar] [CrossRef] [Green Version]
- Banko, P.; Lee, S.Y.; Nagygyorgy, V.; Zrinyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Yao, L.; Tang, Y.; Jhong, J.H.; Wan, J.; Chang, J.; Cui, S.; Luo, Y.; Cai, X.; Li, W.; et al. CircNet 2.0: An updated database for exploring circular RNA regulatory networks in cancers. Nucleic Acids Res. 2022, 50, D93–D101. [Google Scholar] [CrossRef]
- Li, Z.; Chen, Z.; Hu, G.; Jiang, Y. Roles of circular RNA in breast cancer: Present and future. Am. J. Transl. Res. 2019, 11, 3945–3954. [Google Scholar]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource; FDA: Silver Spring, Ml, USA, 2016. [Google Scholar]
- Kessler, L.G.; Barnhart, H.X.; Buckler, A.J.; Choudhury, K.R.; Kondratovich, M.V.; Toledano, A.; Guimaraes, A.R.; Filice, R.; Zhang, Z.; Sullivan, D.C.; et al. The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions. Stat. Methods Med. Res. 2015, 24, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.L.; Allen, C.; Henson, D.E. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer 1989, 63, 181–187. [Google Scholar] [CrossRef]
- Bickel, H.; Pinker-Domenig, K.; Bogner, W.; Spick, C.; Bago-Horvath, Z.; Weber, M.; Helbich, T.; Baltzer, P. Quantitative apparent diffusion coefficient as a noninvasive imaging biomarker for the differentiation of invasive breast cancer and ductal carcinoma in situ. Investig. Radiol. 2015, 50, 95–100. [Google Scholar] [CrossRef] [Green Version]
- Sickles, E. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Shukla-Dave, A.; Obuchowski, N.A.; Chenevert, T.L.; Jambawalikar, S.; Schwartz, L.H.; Malyarenko, D.; Huang, W.; Noworolski, S.M.; Young, R.J.; Shiroishi, M.S.; et al. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials. J. Magn. Reson. Imaging 2019, 49, e101–e121. [Google Scholar] [CrossRef]
- Rogers, W.; Thulasi Seetha, S.; Refaee, T.A.G.; Lieverse, R.I.Y.; Granzier, R.W.Y.; Ibrahim, A.; Keek, S.A.; Sanduleanu, S.; Primakov, S.P.; Beuque, M.P.L.; et al. Radiomics: From qualitative to quantitative imaging. Br. J. Radiol. 2020, 93, 20190948. [Google Scholar] [CrossRef]
- Graham, M.M.; Peterson, L.M.; Hayward, R.M. Comparison of simplified quantitative analyses of FDG uptake. Nucl. Med. Biol. 2000, 27, 647–655. [Google Scholar] [CrossRef]
- Baltzer, P.; Mann, R.M.; Iima, M.; Sigmund, E.E.; Clauser, P.; Gilbert, F.J.; Martincich, L.; Partridge, S.C.; Patterson, A.; Pinker, K.; et al. Diffusion-weighted imaging of the breast-a consensus and mission statement from the EUSOBI International Breast Diffusion-Weighted Imaging working group. Eur. Radiol. 2020, 30, 1436–1450. [Google Scholar] [CrossRef] [Green Version]
- Incoronato, M.; Grimaldi, A.M.; Cavaliere, C.; Inglese, M.; Mirabelli, P.; Monti, S.; Ferbo, U.; Nicolai, E.; Soricelli, A.; Catalano, O.A.; et al. Relationship between functional imaging and immunohistochemical markers and prediction of breast cancer subtype: A PET/MRI study. Eur. J. Nucl. Med. Mo.l Imaging 2018, 45, 1680–1693. [Google Scholar] [CrossRef]
- Castiglioni, I.; Rundo, L.; Codari, M.; Di Leo, G.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI applications to medical images: From machine learning to deep learning. Phys. Med. 2021, 83, 9–24. [Google Scholar] [CrossRef]
- Incoronato, M.; Aiello, M.; Infante, T.; Cavaliere, C.; Grimaldi, A.M.; Mirabelli, P.; Monti, S.; Salvatore, M. Radiogenomic Analysis of Oncological Data: A Technical Survey. Int. J. Mol. Sci. 2017, 18, 805. [Google Scholar] [CrossRef] [Green Version]
- Foster, B.; Bagci, U.; Mansoor, A.; Xu, Z.; Mollura, D.J. A review on segmentation of positron emission tomography images. Comput. Biol. Med. 2014, 50, 76–96. [Google Scholar] [CrossRef]
- Gallivanone, F.; Panzeri, M.M.; Canevari, C.; Losio, C.; Gianolli, L.; De Cobelli, F.; Castiglioni, I. Biomarkers from in vivo molecular imaging of breast cancer: Pretreatment (18)F-FDG PET predicts patient prognosis, and pretreatment DWI-MR predicts response to neoadjuvant chemotherapy. MAGMA 2017, 30, 359–373. [Google Scholar] [CrossRef] [Green Version]
- Veeraraghavan, H.; Dashevsky, B.Z.; Onishi, N.; Sadinski, M.; Morris, E.; Deasy, J.O.; Sutton, E.J. Appearance Constrained Semi-Automatic Segmentation from DCE-MRI is Reproducible and Feasible for Breast Cancer Radiomics: A Feasibility Study. Sci. Rep. 2018, 8, 4838. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Sun, W.; Tseng, T.B.; Li, C.; Qian, W. Fast and fully-automated detection and segmentation of pulmonary nodules in thoracic CT scans using deep convolutional neural networks. Comput. Med. Imaging Graph. 2019, 74, 25–36. [Google Scholar] [CrossRef]
- Leijenaar, R.T.; Carvalho, S.; Velazquez, E.R.; van Elmpt, W.J.; Parmar, C.; Hoekstra, O.S.; Hoekstra, C.J.; Boellaard, R.; Dekker, A.L.; Gillies, R.J.; et al. Stability of FDG-PET Radiomics features: An integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013, 52, 1391–1397. [Google Scholar] [CrossRef] [Green Version]
- Gallivanone, F.; D’Ambrosio, D.; Carne, I.; D’Arcangelo, M.; Montagna, P.; Giroletti, E.; Poggi, P.; Vellani, C.; Moro, L.; Castiglioni, I. A tri-modal tissue-equivalent anthropomorphic phantom for PET, CT and multi-parametric MRI radiomics. Phys. Med. 2022, 98, 28–39. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallieres, M.; Abdalah, M.A.; Aerts, H.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Sardanelli, F.; Aase, H.S.; Alvarez, M.; Azavedo, E.; Baarslag, H.J.; Balleyguier, C.; Baltzer, P.A.; Beslagic, V.; Bick, U.; Bogdanovic-Stojanovic, D.; et al. Position paper on screening for breast cancer by the European Society of Breast Imaging (EUSOBI) and 30 national breast radiology bodies from Austria, Belgium, Bosnia and Herzegovina, Bulgaria, Croatia, Czech Republic, Denmark, Estonia, Finland, France, Germany, Greece, Hungary, Iceland, Ireland, Italy, Israel, Lithuania, Moldova, The Netherlands, Norway, Poland, Portugal, Romania, Serbia, Slovakia, Spain, Sweden, Switzerland and Turkey. Eur. Radiol. 2017, 27, 2737–2743. [Google Scholar]
- Nazari, S.S.; Mukherjee, P. An overview of mammographic density and its association with breast cancer. Breast Cancer 2018, 25, 259–267. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Fishell, E.; Wright, B.; Hanna, W.; Allan, S.; Boyd, N.F. Case-control study of factors associated with failure to detect breast cancer by mammography. J. Natl. Cancer Inst. 1992, 84, 781–785. [Google Scholar] [CrossRef]
- Kim, M.Y.; Choi, N.; Yang, J.H.; Yoo, Y.B.; Park, K.S. Background parenchymal enhancement on breast MRI and mammographic breast density: Correlation with tumour characteristics. Clin. Radiol. 2015, 70, 706–710. [Google Scholar] [CrossRef]
- Wang, S.; Sun, Y.; Mao, N.; Duan, S.; Li, Q.; Li, R.; Jiang, T.; Wang, Z.; Xie, H.; Gu, Y. Incorporating the clinical and radiomics features of contrast-enhanced mammography to classify breast lesions: A retrospective study. Quant. Imaging Med. Surg. 2021, 11, 4418–4430. [Google Scholar] [CrossRef]
- Lin, F.; Wang, Z.; Zhang, K.; Yang, P.; Ma, H.; Shi, Y.; Liu, M.; Wang, Q.; Cui, J.; Mao, N.; et al. Contrast-Enhanced Spectral Mammography-Based Radiomics Nomogram for Identifying Benign and Malignant Breast Lesions of Sub-1 cm. Front. Oncol. 2020, 10, 573630. [Google Scholar] [CrossRef]
- Son, J.; Lee, S.E.; Kim, E.K.; Kim, S. Prediction of breast cancer molecular subtypes using radiomics signatures of synthetic mammography from digital breast tomosynthesis. Sci. Rep. 2020, 10, 21566. [Google Scholar] [CrossRef]
- Qiu, X.; Jiang, Y.; Zhao, Q.; Yan, C.; Huang, M.; Jiang, T. Could Ultrasound-Based Radiomics Noninvasively Predict Axillary Lymph Node Metastasis in Breast Cancer? J. Ultrasound Med. 2020, 39, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Liu, Z.; Sun, C.; Zhang, L.; Wang, Y.; Li, Z.; Shi, J.; Wu, T.; Cui, H.; Zhang, J.; et al. Deep learning radiomics of ultrasonography: Identifying the risk of axillary non-sentinel lymph node involvement in primary breast cancer. EBioMedicine 2020, 60, 103018. [Google Scholar] [CrossRef]
- Ma, M.; Gan, L.; Jiang, Y.; Qin, N.; Li, C.; Zhang, Y.; Wang, X. Radiomics Analysis Based on Automatic Image Segmentation of DCE-MRI for Predicting Triple-Negative and Nontriple-Negative Breast Cancer. Comput. Math. Methods Med. 2021, 2021, 2140465. [Google Scholar] [CrossRef]
- Di Micco, R.; Santurro, L.; Gasparri, M.L.; Zuber, V.; Cisternino, G.; Baleri, S.; Morgante, M.; Rotmensz, N.; Canevari, C.; Gallivanone, F.; et al. PET/MRI for Staging the Axilla in Breast Cancer: Current Evidence and the Rationale for SNB vs. PET/MRI Trials. Cancers 2021, 13, 3751. [Google Scholar] [CrossRef]
- Ming, Y.; Wu, N.; Qian, T.; Li, X.; Wan, D.Q.; Li, C.; Li, Y.; Wu, Z.; Wang, X.; Liu, J.; et al. Progress and Future Trends in PET/CT and PET/MRI Molecular Imaging Approaches for Breast Cancer. Front. Oncol. 2020, 10, 1301. [Google Scholar] [CrossRef]
- Umutlu, L.; Kirchner, J.; Bruckmann, N.M.; Morawitz, J.; Antoch, G.; Ingenwerth, M.; Bittner, A.K.; Hoffmann, O.; Haubold, J.; Grueneisen, J.; et al. Multiparametric Integrated (18)F-FDG PET/MRI-Based Radiomics for Breast Cancer Phenotyping and Tumor Decoding. Cancers 2021, 13, 2928. [Google Scholar] [CrossRef] [PubMed]
- Romeo, V.; Clauser, P.; Rasul, S.; Kapetas, P.; Gibbs, P.; Baltzer, P.A.T.; Hacker, M.; Woitek, R.; Helbich, T.H.; Pinker, K. AI-enhanced simultaneous multiparametric (18)F-FDG PET/MRI for accurate breast cancer diagnosis. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Krajnc, D.; Papp, L.; Nakuz, T.S.; Magometschnigg, H.F.; Grahovac, M.; Spielvogel, C.P.; Ecsedi, B.; Bago-Horvath, Z.; Haug, A.; Karanikas, G.; et al. Breast Tumor Characterization Using [(18)F]FDG-PET/CT Imaging Combined with Data Preprocessing and Radiomics. Cancers 2021, 13, 1249. [Google Scholar] [CrossRef] [PubMed]
- Fantini, L.; Belli, M.L.; Azzali, I.; Loi, E.; Bettinelli, A.; Feliciani, G.; Mezzenga, E.; Fedeli, A.; Asioli, S.; Paganelli, G.; et al. Exploratory Analysis of (18)F-3’-deoxy-3’-fluorothymidine ((18)F-FLT) PET/CT-Based Radiomics for the Early Evaluation of Response to Neoadjuvant Chemotherapy in Patients With Locally Advanced Breast Cancer. Front. Oncol. 2021, 11, 601053. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Zhang, J.; Grimm, L.J.; Yoon, S.C.; Silber, J.I. Radiogenomic analysis of breast cancer: Luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 2014, 273, 365–372. [Google Scholar] [CrossRef]
- Woodard, G.A.; Price, E.R. Qualitative Radiogenomics: Association Between BI-RADS Calcification Descriptors and Recurrence Risk as Assessed by the Oncotype DX Ductal Carcinoma In Situ Score. AJR Am. J. Roentgenol. 2019, 212, 919–924. [Google Scholar] [CrossRef]
- Gallivanone, F.; Cava, C.; Corsi, F.; Bertoli, G.; Castiglioni, I. In Silico Approach for the Definition of radiomiRNomic Signatures for Breast Cancer Differential Diagnosis. Int. J. Mol. Sci. 2019, 20, 5825. [Google Scholar] [CrossRef] [Green Version]
- Incoronato, M.; Grimaldi, A.M.; Mirabelli, P.; Cavaliere, C.; Parente, C.A.; Franzese, M.; Staibano, S.; Ilardi, G.; Russo, D.; Soricelli, A.; et al. Circulating miRNAs in Untreated Breast Cancer: An Exploratory Multimodality Morpho-Functional Study. Cancers 2019, 11, 876. [Google Scholar] [CrossRef] [Green Version]
- Yeh, A.C.; Li, H.; Zhu, Y.; Zhang, J.; Khramtsova, G.; Drukker, K.; Edwards, A.; McGregor, S.; Yoshimatsu, T.; Zheng, Y.; et al. Radiogenomics of breast cancer using dynamic contrast enhanced MRI and gene expression profiling. Cancer Imaging 2019, 19, 48. [Google Scholar] [CrossRef] [Green Version]
- Bismeijer, T.; van der Velden, B.H.M.; Canisius, S.; Lips, E.H.; Loo, C.E.; Viergever, M.A.; Wesseling, J.; Gilhuijs, K.G.A.; Wessels, L.F.A. Radiogenomic Analysis of Breast Cancer by Linking MRI Phenotypes with Tumor Gene Expression. Radiology 2020, 296, 277–287. [Google Scholar] [CrossRef]
- Arefan, D.; Hausler, R.M.; Sumkin, J.H.; Sun, M.; Wu, S. Predicting cell invasion in breast tumor microenvironment from radiological imaging phenotypes. BMC Cancer 2021, 21, 370. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, H.; Guo, W.; Drukker, K.; Lan, L.; Giger, M.L.; Ji, Y. Deciphering Genomic Underpinnings of Quantitative MRI-based Radiomic Phenotypes of Invasive Breast Carcinoma. Sci. Rep. 2015, 5, 17787. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, S.; Han, W.; Kim, Y.; Du, L.; Jamshidi, N.; Huang, D.; Kim, J.H.; Kuo, M.D. Breast Cancer: Radiogenomic Biomarker Reveals Associations among Dynamic Contrast-enhanced MR Imaging, Long Noncoding RNA, and Metastasis. Radiology 2015, 275, 384–392. [Google Scholar] [CrossRef]
- Woodard, G.A.; Ray, K.M.; Joe, B.N.; Price, E.R. Qualitative Radiogenomics: Association between Oncotype DX Test Recurrence Score and BI-RADS Mammographic and Breast MR Imaging Features. Radiology 2018, 286, 60–70. [Google Scholar] [CrossRef] [Green Version]
- Lam, S.W.; Jimenez, C.R.; Boven, E. Breast cancer classification by proteomic technologies: Current state of knowledge. Cancer Treat. Rev. 2014, 40, 129–138. [Google Scholar] [CrossRef]
- Kyndi, M.; Sorensen, F.B.; Knudsen, H.; Overgaard, M.; Nielsen, H.M.; Overgaard, J. Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: The Danish Breast Cancer Cooperative Group. J. Clin. Oncol. 2008, 26, 1419–1426. [Google Scholar] [CrossRef]
- O’Connor, J.P.; Jackson, A.; Parker, G.J.; Roberts, C.; Jayson, G.C. Dynamic contrast-enhanced MRI in clinical trials of antivascular therapies. Nat. Rev. Clin. Oncol. 2012, 9, 167–177. [Google Scholar] [CrossRef]
- Antunovic, L.; Gallivanone, F.; Sollini, M.; Sagona, A.; Invento, A.; Manfrinato, G.; Kirienko, M.; Tinterri, C.; Chiti, A.; Castiglioni, I. [(18)F]FDG PET/CT features for the molecular characterization of primary breast tumors. Eur J. Nucl Med. Mol. Imaging 2017, 44, 1945–1954. [Google Scholar] [CrossRef]
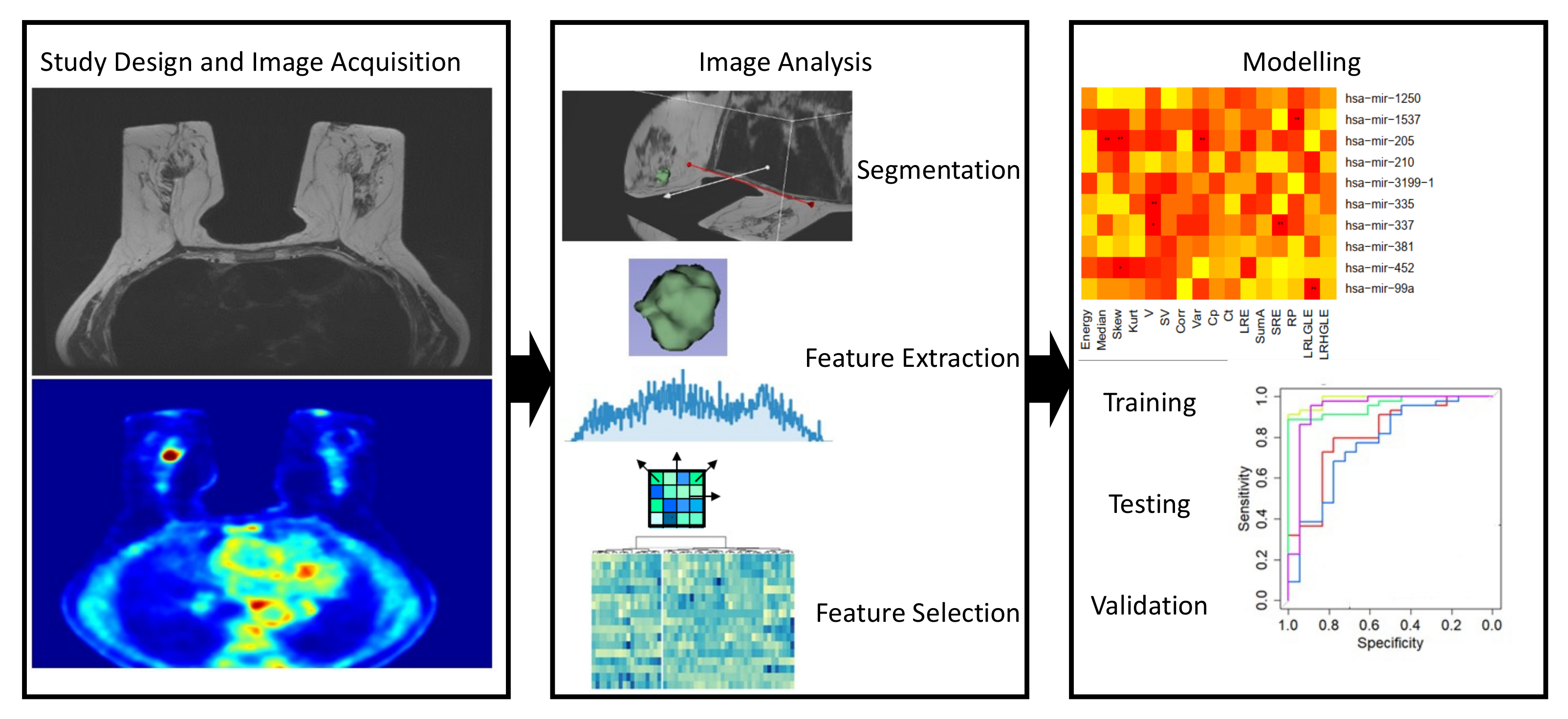

| Imaging | Aim | BC Patients | Data Source | IFs | Findings | Ref. |
|---|---|---|---|---|---|---|
| DCE-MRI | molecular subtype (determined on the basis of genomic analysis) vs. IFs | N = 48 | TCIA-TCGA | Morphological IFs, Local intensity IFs (from kinetics) GLCM IFs | There is an association between dynamic contrast material–enhancement IF that quantifies the relationship between lesion enhancement and background parenchymal enhancement and luminal B subtype | [67] |
| Mammography and MRI | IFs vs. Oncotype DX Test Recurrence Score | N = 408 | Retrospective in-house clinical protocol | Semantic IFs | Semantic IFs from mammography and MRI can be used for imaging biomarkers of breast cancer recurrence risk | [68] |
| DCE-MRI | IFs vs. miRNAs, mRNAs, and regulatory networks | N = 37 | TCIA-TCGA | Morphological IFs, Histogram intensity IFs, GLCM IFs, GLRLM IFs | A radiomiRNomic signature including both miRNAs and imaging features have better classification power of Luminal A versus the different BC subtypes than using miRNAs or imaging alone | [69] |
| PET/MRI | IFs vs. circulating miRNAs | N = 77 | Prospective in-house clinical protocol | Morphological and Local intensity IFs | Different Local intensity IFs have a correlation with miRNAs expression, showing potential for risk stratification of BC and to improve diagnostic accuracy | [70] |
| DCE-MRI | IFs vs. and RNA genomic profile | N = 47 | Retrospective in-house clinical protocol | Morphological IFs, Local intensity IFs (from kinetics) GLCM IFs, | Several molecular pathways related to replication, proliferation, apoptosis, immune system regulation and extracellular signalling have a robust association to IFs | [71] |
| DCE-MRI | IFs vs. gene expression levels from RNA sequencing | N = 295 | Prospective in-house clinical protocol | Morphological IFs, Local intensity IFs (from kinetics) | DCE-MRI phenotypes are related to underlying molecular biology revealed by using RNA sequencing | [72] |
| DCE-MRI | IFs for prediction of cell invasion in the tumor microenvironment | N = 73 | TCIA-TCGA | Morphological IFs, Histogram intensity IFs, GLCM IFs, GLRLM IFs, GLSZM IFs, | Univariate correlations of IFs and abundance of fibroblasts. Multivariate models with AUCs ranging from 0.5 to 0.68 for the multiple cell type invasion predictions | [73] |
| DCE-MRI | IFs vs. DNA mutation, miRNA expression, protein expression, pathway gene expression and copy number variation | N = 91 | TCIA-TCGA | Morphological IFs, Local intensity IFs (from kinetics) GLCM IFs | MRI is a potential non-invasive approach to probe the cancer molecular status, since several transcriptional activities of various genetic pathways were positively associated with different IFs | [74] |
| DCE-MRI | IFs vs. lncRNA expression and MFS | N = 70 | Morphological IFs, Local intensity IFs (from kinetics), Histogram intensity IFs | 5 lncRNAs, involved in the control of cell cycle, cell survival or apoptosis, cellular development, and cell growth, are associated with IFs | [75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallivanone, F.; Bertoli, G.; Porro, D. Radiogenomics, Breast Cancer Diagnosis and Characterization: Current Status and Future Directions. Methods Protoc. 2022, 5, 78. https://doi.org/10.3390/mps5050078
Gallivanone F, Bertoli G, Porro D. Radiogenomics, Breast Cancer Diagnosis and Characterization: Current Status and Future Directions. Methods and Protocols. 2022; 5(5):78. https://doi.org/10.3390/mps5050078
Chicago/Turabian StyleGallivanone, Francesca, Gloria Bertoli, and Danilo Porro. 2022. "Radiogenomics, Breast Cancer Diagnosis and Characterization: Current Status and Future Directions" Methods and Protocols 5, no. 5: 78. https://doi.org/10.3390/mps5050078
APA StyleGallivanone, F., Bertoli, G., & Porro, D. (2022). Radiogenomics, Breast Cancer Diagnosis and Characterization: Current Status and Future Directions. Methods and Protocols, 5(5), 78. https://doi.org/10.3390/mps5050078






