Comparison and Validation of Ichthyoplankton DNA Extraction Methods
Abstract
1. Introduction
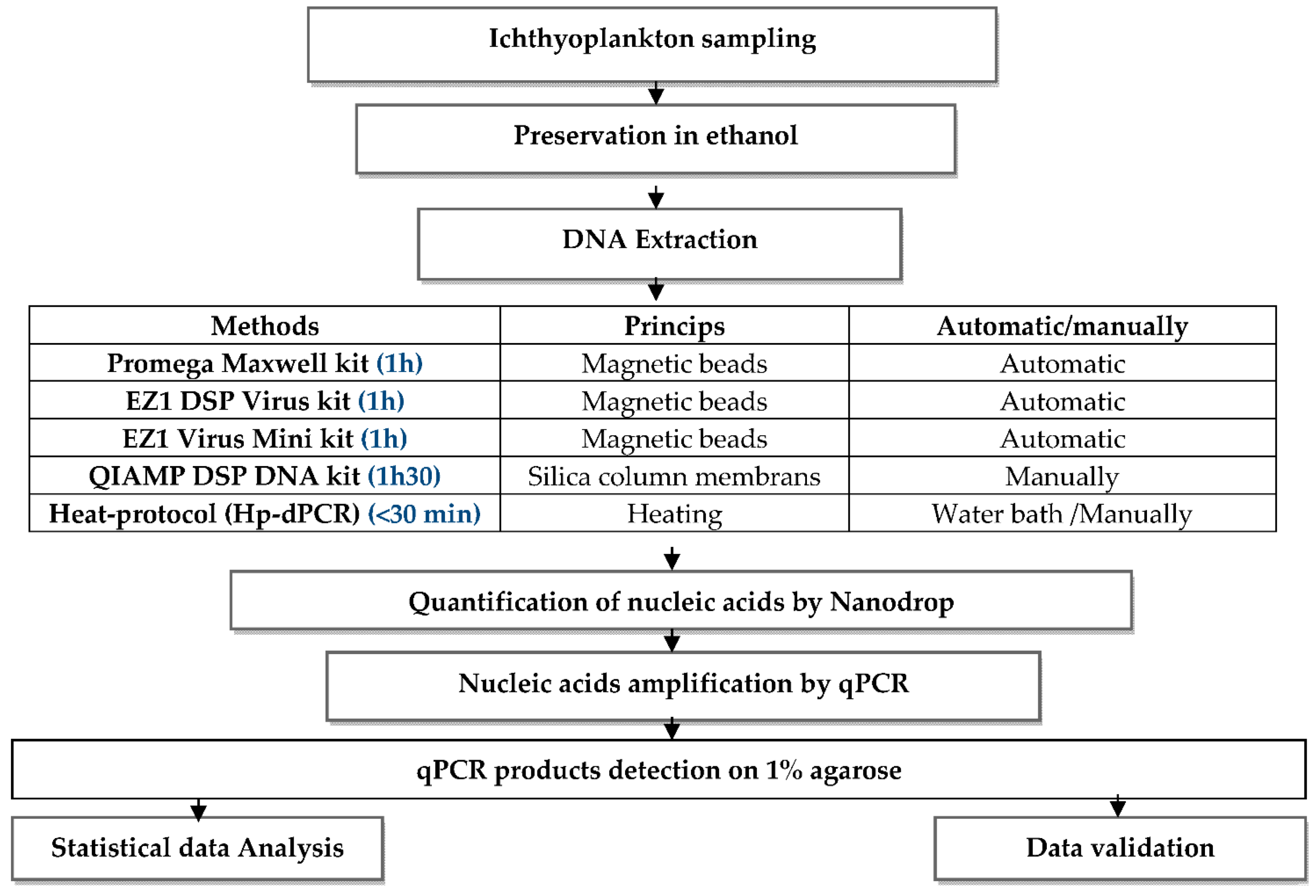
2. Materials and Methods
- I.
- Ichthyoplankton sampling
- II.
- Extraction of nucleic acid
- 1.
- Extraction with EZ1 DSP Virus Kit (Qiagen, Hilden, Germany) and EZ1 Virus Mini Kit (Qiagen, Hilden, Germany)
- 2.
- Extraction with the QIAamp DSP DNA Kit (Qiagen, Hilden, Germany)
- 3.
- Extraction with the Promega Maxwell Kit (Promega Corporation, Madison, WI, USA)
- 4.
- Heat-protocol for direct PCR (Hp-dPCR) (Our protocol)
- III.
- Quantification of nucleic acids
- IV.
- Amplification of nucleic acids
- V.
- Statistical analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Diouri, L.; Soukri, A.; Abdelouahab, H.; Malki, M.; Baibai, T. The Moroccan Ichtyoplankton-A General Overview. Annu. Res. Rev. Biol. 2020, 35, 89–94. [Google Scholar]
- Sedletskaya, V.A. Manuel D’identification des Œufs et des Larves des Espèces les Plus Abondantes de Poissons Habitant au Large des Côtes de L’afrique du Nord-Est; CNROP/AtlantNIRO: Kaliningrad, Russia, 1999. [Google Scholar]
- Taylor, M.I.; Fox, C.; Rico, I.; Rico, C. Species-specific TaqMan probes for simultaneous identification of (Gadus morhua L.), haddock (Melanogrammus aeglefinus L.) and whiting (Merlangius merlangus L.). Mol. Ecol. Notes 2002, 2, 599–601. [Google Scholar] [CrossRef]
- Pappalardo, A.M.; Cuttitta, A.; Sardella, A.; Musco, M.; Maggio, T.; Patti, B.; Mazzola, S.; Ferrito, V. DNA barcoding and COI sequence variation in Mediterranean lanternfishes larvae. Hydrobiologia 2015, 749, 155–167. [Google Scholar] [CrossRef]
- Caldarelli-Stefano, R.; Vago, L.; Bonetto, S.; Nebuloni, M.; Costanzi, G. Use of magnetic beads for tissue DNA extraction and IS6110 Mycobacterium tuberculosis PCR. Mol. Pathol. 1999, 52, 158–160. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Quinteiro, J.; Sotelo, C.G.; Rehbein, H.; Pryde, S.E.; Medina, I.; Perez-Martın, R.I.; Rey-Mendez, M.; Mackie, I.M. Use of mtDNA Direct Polymerase Chain Reaction (PCR) Sequencing and PCR-Restriction Fragment Length Polymorphism Methodologies in Species Identification of Canned Tuna. J. Agric. Food Chem. 1998, 46, 1662–1669. [Google Scholar] [CrossRef]
- Jérôme, M.; Lemaire, C.; Verrez-Bagnis, V.; Etienne, M. Direct Sequencing Method for Species Identification of Canned Sardine and Sardine-Type Products. J. Agric. Food Chem. 2003, 51, 7326–7332. [Google Scholar] [CrossRef] [PubMed]
- Lelievre, S.; Verrez-Bagnis, V.; Jerome, M.; Vaz, S. PCR-RFLP analyses of formalin-fixed fish eggs for the mapping of spawning areas in the Eastern Channel and Southern North Sea. J. Plankton Res. 2010, 32, 1527–1539. [Google Scholar] [CrossRef]
- Berraho, A.; Ettahiri, O.; Letourneur, Y.; Orbi, A.; Yahyaoui, A. Importance des paramètres hydrologiques dans la distribution des œufs et des larves des petits pélagiques du sud de l’Atlantique marocain. Cybium 2005, 29, 21–31. [Google Scholar]
- Ward, R.D.; Hanner, R.; Hebert, P.D.N. The campaign to DNA barcode all fishes, FISH-BOL. J. Fish Biol. 2009, 74, 329–356. [Google Scholar] [CrossRef] [PubMed]
- Larson, J.H.; McCalla, S.G.; Chapman, D.C.; Rees, C.; Knights, B.C.; Vallazza, J.M.; George, A.E.; Richardson, W.B.; Amberg, J. Genetic Analysis Shows that Morphology Alone Cannot Distinguish Asian Carp Eggs from Those of Other Cyprinid Species. N. Am. J. Fish. Manag. 2016, 36, 1053–1058. [Google Scholar] [CrossRef]
- Triant, D.A.; Whitehead, A. Simultaneous Extraction of High-Quality RNA and DNA from Small Tissue Samples. J. Hered. 2009, 100, 246–250. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz-Ramos, J.M.; Hernández-Triana, L.M.; García-De la Peña, C.; González-Álvarez, V.H.; Weger-Lucarelli, J.; Siller-Rodríguez, Q.K.; Sánchez Rámos, F.J.; Rodríguez, A.D.; Ortega-Morales, A.I. Comparison of two DNA extraction methods from larvae, pupae, and adults of Aedes aegypti. Heliyon 2019, 5, e02660. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Guo, S.; Song, N.; Zhang, X. Identification of Cynoglossus joyneri eggs and larvae by DNA barcoding and morphological method. Biodivers. Sci. 2017, 25, 847–855. [Google Scholar] [CrossRef]
- Aranishi, F. Single fish egg DNA extraction for PCR amplification. Conserv. Genet. 2006, 7, 153–156. [Google Scholar] [CrossRef]
- Olekšáková, T.; Žurovcová, M.; Klimešová, V.; Barták, M.; Šuláková, H. DNA extraction and barcode identification of development stages of forensically important flies in the Czech Republic. Mitochondrial DNA Part A 2018, 29, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Becker, R.A.; Sales, N.G.; Santos, G.M.; Santos, G.B.; Carvalho, D.C. DNA barcoding and morphological identification of neotropical ichthyoplankton from the Upper Paraná and São Francisco: DNA barcoding of neotropical ichthyoplankton. J. Fish Biol. 2015, 87, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Kerr, M.; Browning, J.; Bønnelycke, E.; Zhang, Y.; Hu, C.; Armenteros, M.; Murawski, S.; Peebles, E.; Breitbart, M. DNA barcoding of fish eggs collected off northwestern Cuba and across the Florida Straits demonstrates egg transport by mesoscale eddies. Fish. Oceanogr. 2020, 29, 340–348. [Google Scholar] [CrossRef]
- Gleason, L.U.; Burton, R.S. High-throughput molecular identification of fish eggs using multiplex suspension bead arrays. Mol. Ecol. Resour. 2012, 12, 57–66. [Google Scholar] [CrossRef] [PubMed]

| Extraction Method | Concentration ng/µL | R 260/280 |
|---|---|---|
| EZ1 DSP Virus Kit (Qiagen, Hilden, Germany) | 0.8 ± 0.329 | 1.5 ± 0.086 |
| EZ1 Virus Mini Kit (Qiagen, Hilden, Germany) | 7 ± 0.243 | 1.3 ± 0.256 |
| QIAmp DSP DNA Kit (Qiagen, Hilden, Germany) | 3 ± 0.598 | 1.6 ± 0.092 |
| Promega Maxwell Kit (Promega Corporation, Madison, WI, USA) | 50 ± 0.053 | 1.7 ± 0.624 |
| Heat-protocol for direct PCR (Hp-dPCR) (Our protocol) | 170 ± 0.717 | 1.8 ± 0.044 |
| Kits | Costs/Kit | Costs/Test | Extraction Duration | Required Instruments |
|---|---|---|---|---|
| EZ1 DSP Virus Kit | * 475.00$/48 tests | 9.89$ | 1 h | EZ1 advanced XL |
| EZ1 Virus Kit | * 443.00$/48 tests | 9.23$ | 1 h | EZ1 advanced XL |
| QIAamp DNA Kit | * 259.00$/50 tests | 5.18$ | 1 h | QIAcube |
| Promega Maxwell Kit | * 750.00$/48test | 15.63$ | 1 h 30 min | Maxwell® RSC |
| Hp-dPCR Method | Not on the market 3.00$/test | 3.00$ | <30 min | Water bath |
| Extraction Method | Ct (Cycle Threshold) |
|---|---|
| EZ1 DSP Virus Kit (Qiagen, Hilden, Germany) | ------- |
| EZ1 Virus Mini Kit (Qiagen, Hilden, Germany) | ------- |
| QIAamp DSP DNA Kit(Qiagen, Hilden, Germany) | 33.87 ± 0.746 |
| Promega Maxwell Kit (Promega Corporation, Madison, WI, USA) | 20.54 ± 0.4403 |
| Heat-protocol for direct PCR (Hp-dPCR) (Our protocol) | 18.32 ± 0.7043 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamia, D.; Théophile, U.; Hinde, A.; Mohamed, M.; Tarik, B.; Abdelaziz, S. Comparison and Validation of Ichthyoplankton DNA Extraction Methods. Methods Protoc. 2021, 4, 87. https://doi.org/10.3390/mps4040087
Lamia D, Théophile U, Hinde A, Mohamed M, Tarik B, Abdelaziz S. Comparison and Validation of Ichthyoplankton DNA Extraction Methods. Methods and Protocols. 2021; 4(4):87. https://doi.org/10.3390/mps4040087
Chicago/Turabian StyleLamia, Diouri, Uwiringiyeyezu Théophile, Abdelouahab Hinde, Malki Mohamed, Baibai Tarik, and Soukri Abdelaziz. 2021. "Comparison and Validation of Ichthyoplankton DNA Extraction Methods" Methods and Protocols 4, no. 4: 87. https://doi.org/10.3390/mps4040087
APA StyleLamia, D., Théophile, U., Hinde, A., Mohamed, M., Tarik, B., & Abdelaziz, S. (2021). Comparison and Validation of Ichthyoplankton DNA Extraction Methods. Methods and Protocols, 4(4), 87. https://doi.org/10.3390/mps4040087






