Concurrent Measurement of Mitochondrial DNA Copy Number and ATP Concentration in Single Bovine Oocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Cumulus-Oocyte-Complexes
2.2. TaqMan Primer and Probe Design
2.3. Validation
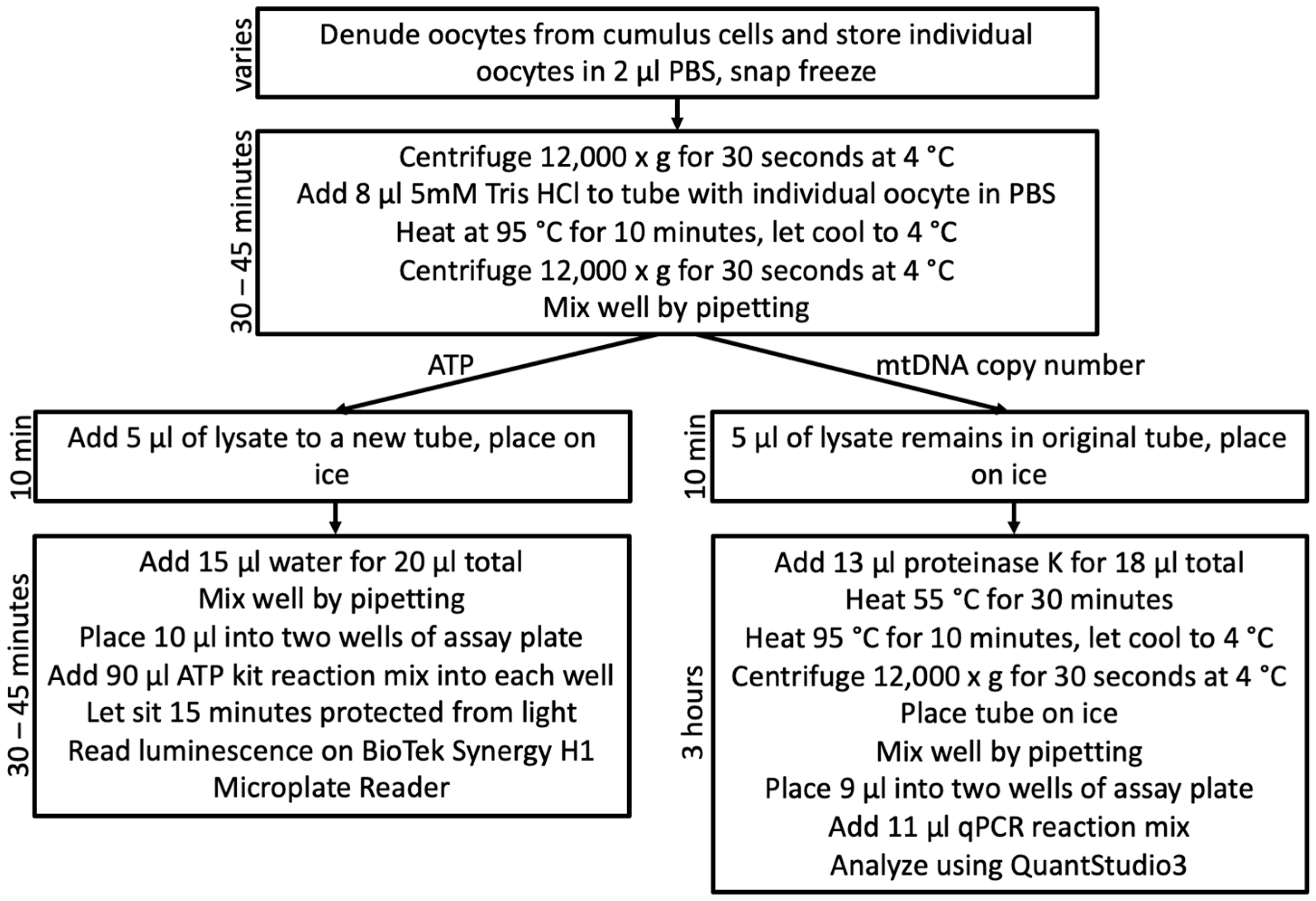
2.3.1. Oocyte Lysis and Division
2.3.2. ATP Quantification Validation
2.3.3. mtDNA Copy Number Quantification Validation
2.4. Germinal Vesicle and Metaphase II Comparison
2.4.1. Oocyte Lysis and Division
2.4.2. ATP Quantification
2.4.3. mtDNA Standard Preparation
2.4.4. mtDNA Copy Number Quantification
2.5. Statistics and Analyses
3. Results and Discussion
3.1. Standards Quality
3.2. Validation of Procedures for Quantification of ATP and mtDNA Copy Number in Quarter Oocyte Equivalents
3.3. ATP and mtDNA Copy Number in GV versus MII Oocytes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stojkovic, M.; Machado, S.A.; Stojkovic, P.; Zakhartchenko, V.; Hutzler, P.; Gonçalves, P.B.; Wolf, E. Mitochondrial distribution and adenosine triphosphate content of bovine oocytes before and after in vitro maturation: Correlation with morphological criteria and developmental capacity after in vitro fertilization and culture. Biol. Reprod. 2001, 64, 904–909. [Google Scholar] [CrossRef] [Green Version]
- Scantland, S.; Tessaro, I.; Macabelli, C.H.; Macaulay, A.D.; Cagnone, G.; Fournier, É.; Luciano, A.M.; Robert, C. The adenosine salvage pathway as an alternative to mitochondrial production of ATP in maturing mammalian oocytes. Biol. Reprod. 2014, 91, 75. [Google Scholar] [CrossRef]
- Iwata, H.; Goto, H.; Tanaka, H.; Sakaguchi, Y.; Kimura, K.; Kuwayama, T.; Monji, Y. Effect of maternal age on mitochondrial DNA copy number, ATP content and IVF outcome of bovine oocytes. Reprod. Fertil. Dev. 2011, 23, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Chiaratti, M.R.; Bressan, F.F.; Ferreira, C.R.; Caetano, A.R.; Smith, L.C.; Vercesi, A.E.; Meirelles, F.V. Embryo mitochondrial DNA depletion is reversed during early embryogenesis in cattle. Biol. Reprod. 2010, 82, 76–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirillova, A.; Smitz, J.E.J.; Sukhikh, G.T.; Mazunin, I. The role of mitochondria in oocyte maturation. Cells 2021, 10, 2484. [Google Scholar] [CrossRef]
- St John, J. The control of mtDNA replication during differentiation and development. Biochim. Biophys. Acta 2014, 1840, 1345–1354. [Google Scholar] [CrossRef]
- Mao, J.; Whitworth, K.M.; Spate, L.D.; Walters, E.M.; Zhao, J.; Prather, R.S. Regulation of oocyte mitochondrial DNA copy number by follicular fluid, EGF, and neuregulin 1 during in vitro maturation affects embryo development in pigs. Theriogenology 2012, 78, 887–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contreras-Solís, I.; Catalá, M.; Soto-Heras, S.; Roura, M.; Paramio, M.T.; Izquierdo, D. Effect of follicle size on hormonal status of follicular fluid, oocyte ATP content, and in vitro embryo production in prepubertal sheep. Domest. Anim. Endocrinol. 2021, 75, 106582. [Google Scholar] [CrossRef]
- May-Panloup, P.; Boguenet, M.; Hachem, H.E.; Bouet, P.E.; Reynier, P. Embryo and its mitochondria. Antioxidants 2021, 10, 139. [Google Scholar] [CrossRef]
- Kidder, G.M.; Vanderhyden, B.C. Bidirectional communication between oocytes and follicle cells: Ensuring oocyte developmental competence. Can. J. Physiol. Pharmacol. 2010, 88, 399–413. [Google Scholar] [CrossRef] [Green Version]
- Biggers, J.; Whittingham, D.; Donahue, R. The pattern of energy metabolism in the mouse oocyte and zygote. Zoology 1967, 58, 560–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutton-McDowall, M.L.; Gilchrist, R.B.; Thompson, J.G. The pivotal role of glucose metabolism in determining oocyte developmental competence. Reproduction 2010, 139, 685–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalbies-Tran, R.; Cadoret, V.; Desmarchais, A.; Elis, S.; Maillard, V.; Monget, P.; Monniaux, D.; Reynaud, K.; Saint-Dizier, M.; Uzbekova, S. A comparative analysis of oocyte development in mammals. Cells 2020, 9, 1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paczkowski, M.; Silva, E.; Schoolcraft, W.B.; Krisher, R.L. Comparative importance of fatty acid beta-oxidation to nuclear maturation, gene expression, and glucose metabolism in mouse, bovine, and porcine cumulus oocyte complexes. Biol. Reprod. 2013, 88, 111. [Google Scholar] [CrossRef] [PubMed]
- Winterhager, E.; Kidder, G.M. Gap junction connexins in female reproductive organs: Implications for women’s reproductive health. Hum. Reprod. Update 2015, 21, 340–352. [Google Scholar] [CrossRef] [Green Version]
- Richani, D.; Dunning, K.R.; Thompson, J.G.; Gilchrist, R.B. Metabolic co-dependence of the oocyte and cumulus cells: Essential role in determining oocyte developmental competence. Hum. Reprod. Update 2021, 27, 27–47. [Google Scholar] [CrossRef]
- Nagano, M. Acquisition of developmental competence and in vitro growth culture of bovine oocytes. J. Reprod. Dev. 2019, 65, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Roth, Z. Symposium review: Reduction in oocyte developmental competence by stress is associated with alterations in mitochondrial function. J. Dairy Sci. 2018, 101, 3642–3654. [Google Scholar] [CrossRef]
- Dumollard, R.; Duchen, M.; Carroll, J. The Role of Mitochondrial Function in the Oocyte and Embryo. Curr. Top. Dev. Biol. 2007, 77, 21–49. [Google Scholar] [CrossRef]
- Hashimoto, S.; Yamanaka, M.; Yamochi, T.; Iwata, H.; Kawahara-Miki, R.; Inoue, M.; Morimoto, Y. Mitochondrial function in immature bovine oocytes is improved by an increase of cellular cyclic AMP. Sci. Rep. 2019, 9, 5167. [Google Scholar] [CrossRef]
- Van Blerkom, J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion 2011, 11, 797–813. [Google Scholar] [CrossRef] [PubMed]
- GraphPad QuickCalcs Molarity Calculator. Available online: https://www.graphpad.com/quickcalcs/molarityform/ (accessed on 1 June 2021).
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 5957, Adenosine-5′-Triphosphate. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Adenosine-5_-triphosphate (accessed on 24 September 2021).
- QPCR Efficiency Calculator. Available online: https://www.thermofisher.com/us/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/qpcr-efficiency-calculator.html (accessed on 1 June 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 1 June 2021).
- Wang, T.Y.; Wang, L.; Zhang, J.H.; Dong, W.H. A simplified universal genomic DNA extraction protocol suitable for PCR. Genet. Mol. Res. 2011, 10, 519–525. [Google Scholar] [CrossRef]
- Thermo Scientific. Interpretation of Nucleic Acid 260/280 Ratios; Thermo Scientific: Waltham, MA, USA, 2012. [Google Scholar]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roche. Overview of LightCycler Quantification Methods; Roche: Basel, Switzerland, 2000. [Google Scholar]
- AppliedBiosystems. Amplification Efficiency of TaqMan® Gene Expression Assays; AppliedBiosystems: Waltham, MA, USA, 2004. [Google Scholar]
- Hazra, A.; Gogtay, N. Biostatistics Series Module 6: Correlation and Linear Regression. Indian J. Dermatol. 2016, 61, 593–601. [Google Scholar] [CrossRef]
- Jiao, F.; Yan, J.B.; Yang, X.Y.; Li, H.; Wang, Q.; Huang, S.Z.; Zeng, F.; Zeng, Y.T. Effect of oocyte mitochondrial DNA haplotype on bovine somatic cell nuclear transfer efficiency. Mol. Reprod. Dev. 2007, 74, 1278–1286. [Google Scholar] [CrossRef]
- Dadarwal, D.; Dias, F.C.F.; Adams, G.P.; Singh, J. Effect of follicular aging on ATP content and mitochondria distribution in bovine oocytes. Theriogenology 2017, 89, 348–358. [Google Scholar] [CrossRef]
- Tamassia, M.; Nuttinck, F.; May-Panloup, P.; Reynier, P.; Heyman, Y.; Charpigny, G.; Stojkovic, M.; Hiendleder, S.; Renard, J.P.; Chastant-Maillard, S. In vitro embryo production efficiency in cattle and its association with oocyte adenosine triphosphate content, quantity of mitochondrial DNA, and mitochondrial DNA haplogroup. Biol. Reprod. 2004, 71, 697–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payton, R.R.; Rispoli, L.A.; Nagle, K.A.; Gondro, C.; Saxton, A.M.; Voy, B.H.; Edwards, J.L. Mitochondrial-related consequences of heat stress exposure during bovine oocyte maturation persist in early embryo development. J. Reprod. Dev. 2018, 64, 243–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeng, J.Y.; Yeh, T.S.; Lee, J.W.; Lin, S.H.; Fong, T.H.; Hsieh, R.H. Maintenance of mitochondrial DNA copy number and expression are essential for preservation of mitochondrial function and cell growth. J. Cell. Biochem. 2008, 103, 347–357. [Google Scholar] [CrossRef]
- May-Panloup, P.; Chretien, M.F.; Malthiery, Y.; Reynier, P. Mitochondrial DNA in the oocyte and the developing embryo. Curr. Top. Dev. Biol. 2007, 77, 51–83. [Google Scholar] [CrossRef] [PubMed]
- Kimble, K.M.; Dickinson, S.E.; Biase, F.H. Extraction of total RNA from single-oocytes and single-cell mRNA sequencing of swine oocytes. BMC Res. Notes 2018, 11, 155. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence | Product Length | Tm |
|---|---|---|---|
| mtDNA Primer Forward | CCTACAAACGCTCCTTCCAC | 183 | 59 |
| mtDNA Primer Reverse | AGAGAATATAGGGCGGTGATTACT | 183 | 59 |
| TaqMan Probe | TTGTTGGGGGTAGAGCTAAGTTGGT | --- | 64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Read, C.C.; Bhandari, S.; Moorey, S.E. Concurrent Measurement of Mitochondrial DNA Copy Number and ATP Concentration in Single Bovine Oocytes. Methods Protoc. 2021, 4, 88. https://doi.org/10.3390/mps4040088
Read CC, Bhandari S, Moorey SE. Concurrent Measurement of Mitochondrial DNA Copy Number and ATP Concentration in Single Bovine Oocytes. Methods and Protocols. 2021; 4(4):88. https://doi.org/10.3390/mps4040088
Chicago/Turabian StyleRead, Casey C., Sadikshya Bhandari, and Sarah E. Moorey. 2021. "Concurrent Measurement of Mitochondrial DNA Copy Number and ATP Concentration in Single Bovine Oocytes" Methods and Protocols 4, no. 4: 88. https://doi.org/10.3390/mps4040088
APA StyleRead, C. C., Bhandari, S., & Moorey, S. E. (2021). Concurrent Measurement of Mitochondrial DNA Copy Number and ATP Concentration in Single Bovine Oocytes. Methods and Protocols, 4(4), 88. https://doi.org/10.3390/mps4040088






