Pichia pastoris and the Recombinant Human Heterodimeric Amino Acid Transporter 4F2hc-LAT1: From Clone Selection to Pure Protein
Abstract
1. Introduction
2. Materials and Methods
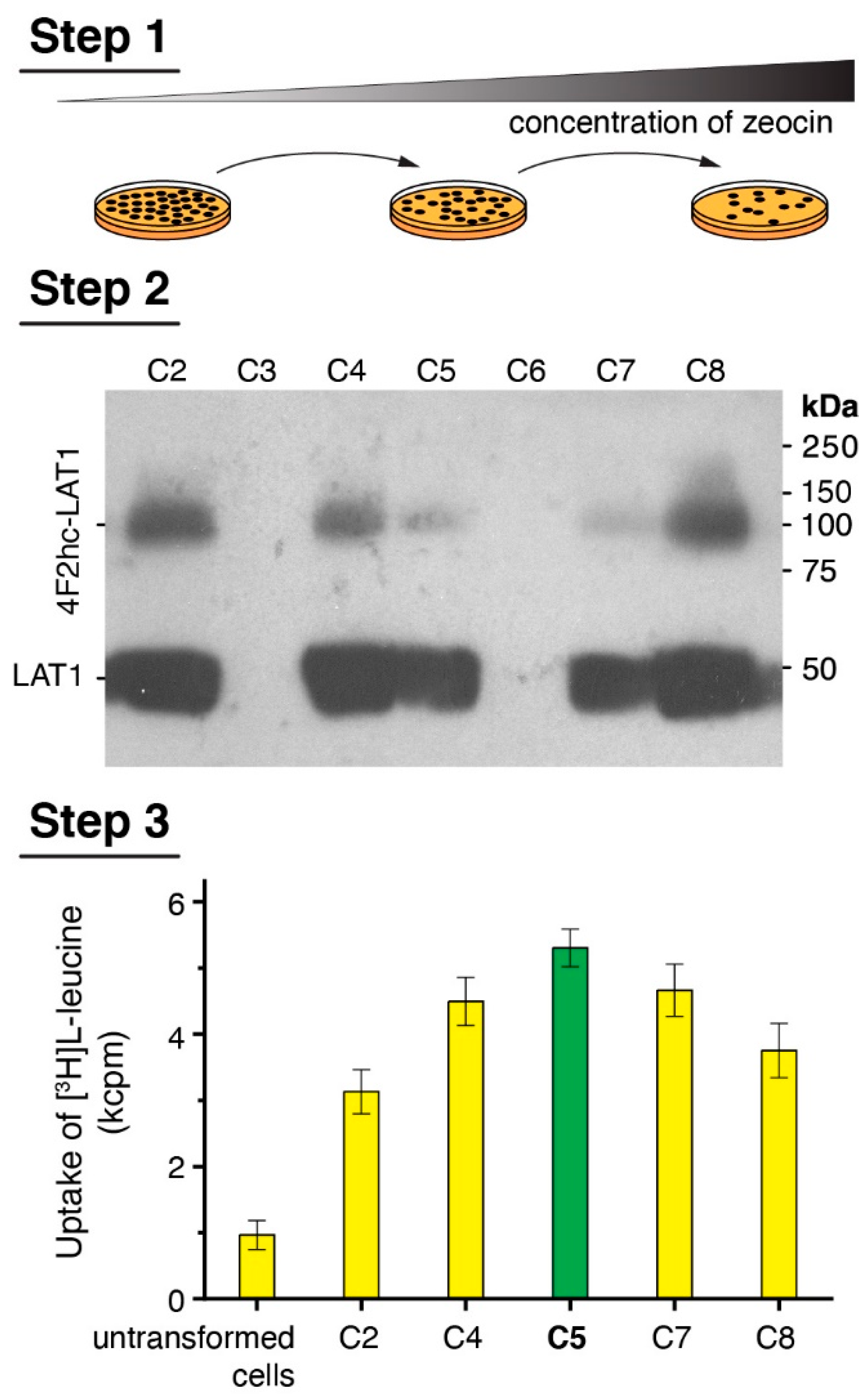
2.1. Clone Picking Strategy
2.2. Large-Scale Overexpression and Membrane Isolation
2.3. Protein Purification
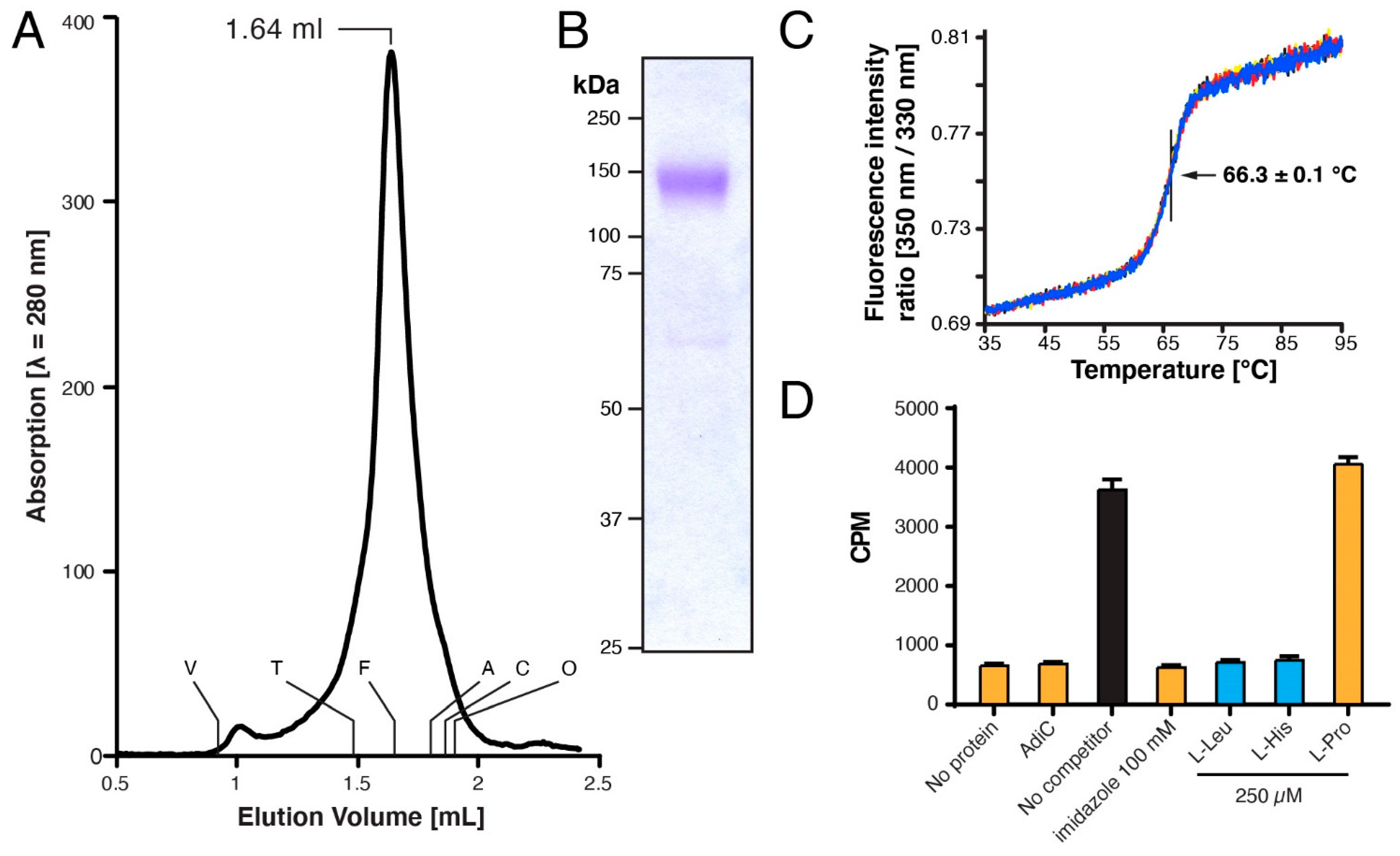
2.4. Size-Exclusion Chromatography
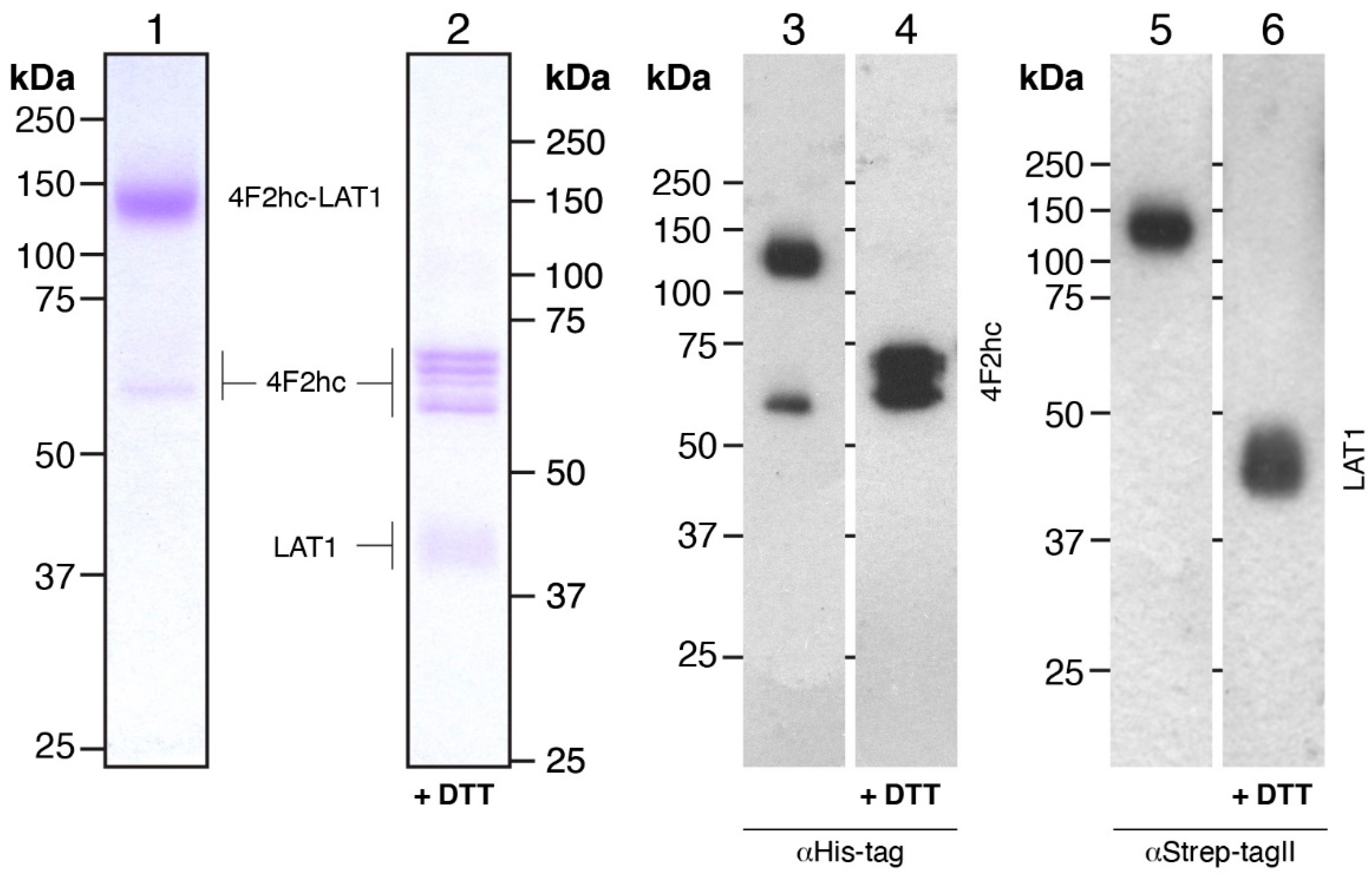
2.5. Gel Electrophoresis and Western Blot Analysis
2.6. Determination of the Thermostability
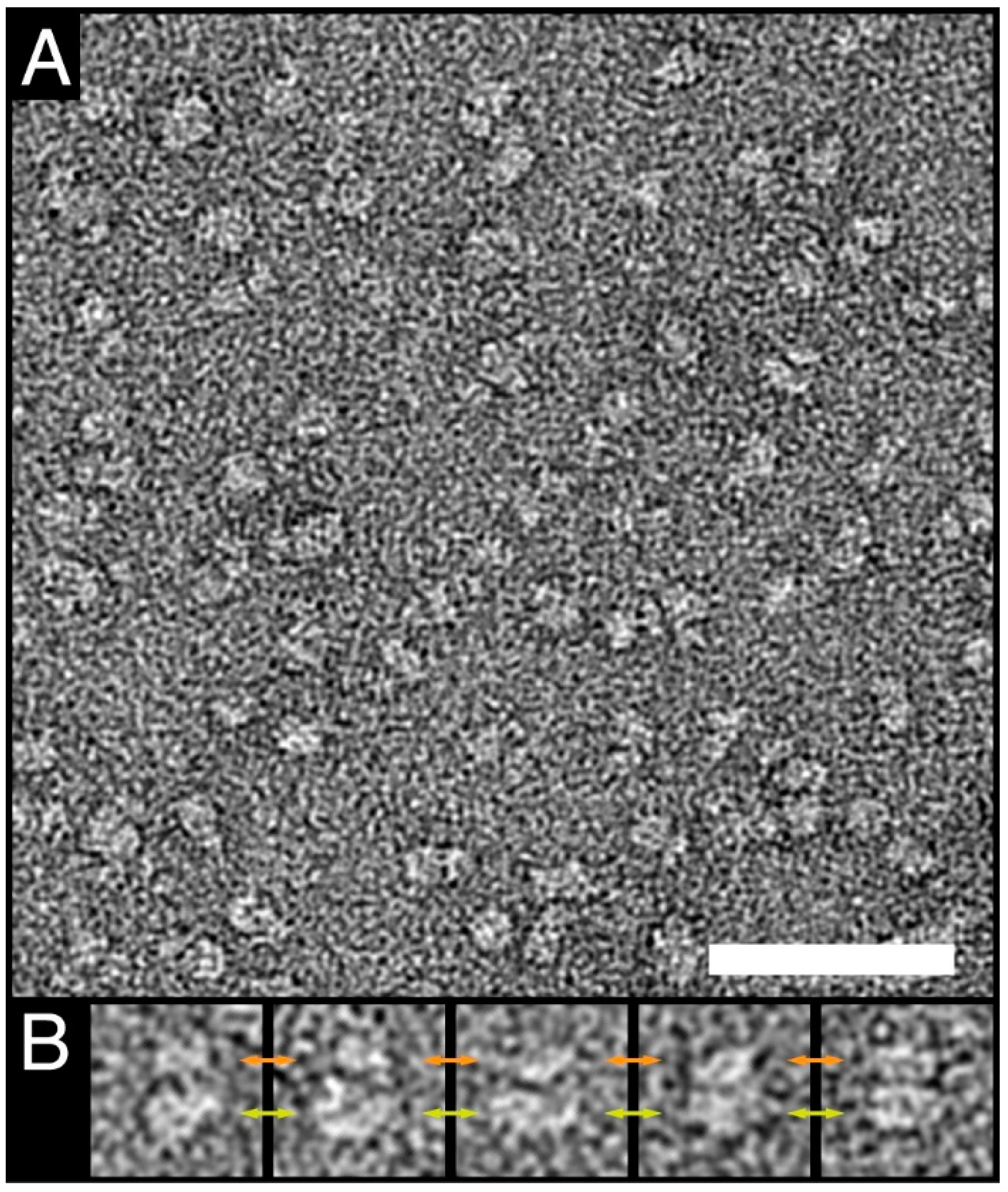
2.7. Negative-Stain Transmission Electron Microscopy
2.8. Scintillation-Proximity Assay (SPA) Analysis
3. Results and Discussion
3.1. Clone Selection Procedure
3.2. Large-Scale Expression and Purification
3.3. Characterization of Purified Human 4F2hc-LAT1 Protein
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Broer, S.; Broer, A. Amino acid homeostasis and signalling in mammalian cells and organisms. Biochem. J. 2017, 474, 1935–1963. [Google Scholar] [CrossRef] [PubMed]
- Broer, S. Amino acid transporters as disease modifiers and drug targets. SLAS Discov. 2018, 23, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Fotiadis, D.; Kanai, Y.; Palacin, M. The SLC3 and SLC7 families of amino acid transporters. Mol. Asp. Med. 2013, 34, 139–158. [Google Scholar] [CrossRef]
- Chillaron, J.; Roca, R.; Valencia, A.; Zorzano, A.; Palacin, M. Heteromeric amino acid transporters: Biochemistry, genetics, and physiology. Am. J. Physiol. Renal Physiol. 2001, 281, F995–F1018. [Google Scholar] [CrossRef]
- Wagner, C.A.; Lang, F.; Broer, S. Function and structure of heterodimeric amino acid transporters. Am. J. Physiol. Cell Physiol. 2001, 281, C1077–C1093. [Google Scholar] [CrossRef]
- Palacin, M.; Kanai, Y. The ancillary proteins of HATs: SLC3 family of amino acid transporters. Pflug. Arch. 2004, 447, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Verrey, F.; Closs, E.I.; Wagner, C.A.; Palacin, M.; Endou, H.; Kanai, Y. CATs and HATs: The SLC7 family of amino acid transporters. Pflug. Arch. 2004, 447, 532–542. [Google Scholar] [CrossRef]
- Napolitano, L.; Scalise, M.; Galluccio, M.; Pochini, L.; Albanese, L.M.; Indiveri, C. LAT1 is the transport competent unit of the LAT1/CD98 heterodimeric amino acid transporter. Int. J. Biochem. Cell Biol. 2015, 67, 25–33. [Google Scholar] [CrossRef]
- Reig, N.; Chillaron, J.; Bartoccioni, P.; Fernandez, E.; Bendahan, A.; Zorzano, A.; Kanner, B.; Palacin, M.; Bertran, J. The light subunit of system b(o,+) is fully functional in the absence of the heavy subunit. EMBO J. 2002, 21, 4906–4914. [Google Scholar] [CrossRef]
- Rosell, A.; Meury, M.; Alvarez-Marimon, E.; Costa, M.; Perez-Cano, L.; Zorzano, A.; Fernandez-Recio, J.; Palacin, M.; Fotiadis, D. Structural bases for the interaction and stabilization of the human amino acid transporter LAT2 with its ancillary protein 4F2hc. Proc. Natl. Acad. Sci. USA 2014, 111, 2966–2971. [Google Scholar] [CrossRef]
- Broer, S.; Palacin, M. The role of amino acid transporters in inherited and acquired diseases. Biochem. J. 2011, 436, 193–211. [Google Scholar] [CrossRef]
- Errasti-Murugarren, E.; Palacin, M. Heteromeric amino acid transporters in brain: From physiology to pathology. Neurochem. Res. 2021. [Google Scholar] [CrossRef]
- Fort, J.; de la Ballina, L.R.; Burghardt, H.E.; Ferrer-Costa, C.; Turnay, J.; Ferrer-Orta, C.; Uson, I.; Zorzano, A.; Fernandez-Recio, J.; Orozco, M.; et al. The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane. J. Biol. Chem. 2007, 282, 31444–31452. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhao, X.; Lei, J.; Zhou, Q. Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature 2019, 568, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Wiriyasermkul, P.; Jin, C.; Quan, L.; Ohgaki, R.; Okuda, S.; Kusakizako, T.; Nishizawa, T.; Oda, K.; Ishitani, R.; et al. Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc. Nat. Struct. Mol. Biol. 2019, 26, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Jeckelmann, J.-M.; Fotiadis, D. Sub-nanometer cryo-EM density map of the human heterodimeric amino acid transporter 4F2hc-LAT2. Int. J. Mol. Sci. 2020, 21, 7094. [Google Scholar] [CrossRef] [PubMed]
- Chiduza, G.N.; Johnson, R.M.; Wright, G.S.A.; Antonyuk, S.V.; Muench, S.P.; Hasnain, S.S. LAT1 (SLC7A5) and CD98hc (SLC3A2) complex dynamics revealed by single-particle cryo-EM. Acta Crystallogr. D Struct. Biol. 2019, 75, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Meury, M.; Costa, M.; Harder, D.; Stauffer, M.; Jeckelmann, J.-M.; Bruhlmann, B.; Rosell, A.; Ilgu, H.; Kovar, K.; Palacin, M.; et al. Detergent-induced stabilization and improved 3D map of the human heteromeric amino acid transporter 4F2hc-LAT2. PLoS ONE 2014, 9, e109882. [Google Scholar] [CrossRef]
- Jeckelmann, J.-M.; Fotiadis, D. Volta phase plate cryo-EM structure of the human heterodimeric amino acid transporter 4F2hc-LAT2. Int. J. Mol. Sci. 2019, 20, 931. [Google Scholar] [CrossRef]
- Yan, R.; Zhou, J.; Li, Y.; Lei, J.; Zhou, Q. Structural insight into the substrate recognition and transport mechanism of the human LAT2-4F2hc complex. Cell Discov. 2020, 6, 82. [Google Scholar] [CrossRef]
- Meier, C.; Ristic, Z.; Klauser, S.; Verrey, F. Activation of system L heterodimeric amino acid exchangers by intracellular substrates. EMBO J. 2002, 21, 580–589. [Google Scholar] [CrossRef]
- Cosco, J.; Scalise, M.; Colas, C.; Galluccio, M.; Martini, R.; Rovella, F.; Mazza, T.; Ecker, G.F.; Indiveri, C. ATP modulates SLC7A5 (LAT1) synergistically with cholesterol. Sci. Rep. 2020, 10, 16738. [Google Scholar] [CrossRef]
- Scalise, M.; Galluccio, M.; Console, L.; Pochini, L.; Indiveri, C. The human SLC7A5 (LAT1): The intriguing histidine/large neutral amino acid transporter and its relevance to human health. Front. Chem. 2018, 6, 243. [Google Scholar] [CrossRef]
- Kantipudi, S.; Jeckelmann, J.-M.; Ucurum, Z.; Bosshart, P.D.; Fotiadis, D. The heavy chain 4F2hc modulates the substrate affinity and specificity of the light chains LAT1 and LAT2. Int. J. Mol. Sci. 2020, 21, 7573. [Google Scholar] [CrossRef]
- Kageyama, T.; Nakamura, M.; Matsuo, A.; Yamasaki, Y.; Takakura, Y.; Hashida, M.; Kanai, Y.; Naito, M.; Tsuruo, T.; Minato, N.; et al. The 4F2hc/LAT1 complex transports L-DOPA across the blood-brain barrier. Brain Res. 2000, 879, 115–121. [Google Scholar] [CrossRef]
- Zevenbergen, C.; Meima, M.E.; Lima de Souza, E.C.; Peeters, R.P.; Kinne, A.; Krause, G.; Visser, W.E.; Visser, T.J. Transport of iodothyronines by human L-type amino acid transporters. Endocrinology 2015, 156, 4345–4355. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Friesema, E.C.; Docter, R.; Moerings, E.P.; Verrey, F.; Krenning, E.P.; Hennemann, G.; Visser, T.J. Thyroid hormone transport by the heterodimeric human system L amino acid transporter. Endocrinology 2001, 142, 4339–4348. [Google Scholar] [CrossRef] [PubMed]
- Nicklin, P.; Bergman, P.; Zhang, B.; Triantafellow, E.; Wang, H.; Nyfeler, B.; Yang, H.; Hild, M.; Kung, C.; Wilson, C.; et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009, 136, 521–534. [Google Scholar] [CrossRef]
- Singh, N.; Ecker, G.F. Insights into the structure, function, and ligand discovery of the large neutral amino acid transporter 1, LAT1. Int. J. Mol. Sci. 2018, 19, 1278. [Google Scholar] [CrossRef]
- Kandasamy, P.; Gyimesi, G.; Kanai, Y.; Hediger, M.A. Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 2018, 43, 752–789. [Google Scholar] [CrossRef] [PubMed]
- Hafliger, P.; Charles, R.-P. The L-type amino acid transporter LAT1-an emerging target in cancer. Int. J. Mol. Sci. 2019, 20, 2428. [Google Scholar] [CrossRef] [PubMed]
- Macauley-Patrick, S.; Fazenda, M.L.; McNeil, B.; Harvey, L.M. Heterologous protein production using the Pichia pastoris expression system. Yeast 2005, 22, 249–270. [Google Scholar] [CrossRef] [PubMed]
- Cereghino, J.L.; Cregg, J.M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000, 24, 45–66. [Google Scholar] [CrossRef]
- Cregg, J.M.; Cereghino, J.L.; Shi, J.; Higgins, D.R. Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 2000, 16, 23–52. [Google Scholar] [CrossRef]
- Looser, V.; Bruhlmann, B.; Bumbak, F.; Stenger, C.; Costa, M.; Camattari, A.; Fotiadis, D.; Kovar, K. Cultivation strategies to enhance productivity of Pichia pastoris: A review. Biotechnol. Adv. 2015, 33, 1177–1193. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Fotiadis, D.; Sjovall, S.; Johansson, I.; Hedfalk, K.; Engel, A.; Kjellbom, P. Reconstitution of water channel function of an aquaporin overexpressed and purified from Pichia pastoris. FEBS Lett. 2003, 537, 68–72. [Google Scholar] [CrossRef]
- Byrne, B. Pichia pastoris as an expression host for membrane protein structural biology. Curr. Opin. Struct. Biol. 2015, 32, 9–17. [Google Scholar] [CrossRef]
- Lyons, J.A.; Shahsavar, A.; Paulsen, P.A.; Pedersen, B.P.; Nissen, P. Expression strategies for structural studies of eukaryotic membrane proteins. Curr. Opin. Struct. Biol. 2016, 38, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Suades, A.; Alcaraz, A.; Cruz, E.; Alvarez-Marimon, E.; Whitelegge, J.P.; Manyosa, J.; Cladera, J.; Peralvarez-Marin, A. Structural biology workflow for the expression and characterization of functional human sodium glucose transporter type 1 in Pichia pastoris. Sci. Rep. 2019, 9, 1203. [Google Scholar] [CrossRef]
- Costa, M.; Rosell, A.; Alvarez-Marimon, E.; Zorzano, A.; Fotiadis, D.; Palacin, M. Expression of human heteromeric amino acid transporters in the yeast Pichia pastoris. Protein Expr. Purif. 2013, 87, 35–40. [Google Scholar] [CrossRef]
- Kantipudi, S.; Fotiadis, D. Yeast cell-based transport assay for the functional characterization of human 4F2hc-LAT1 and -LAT2, and LAT1 and LAT2 substrates and inhibitors. Front. Mol. Biosci. 2021, 8, 676854. [Google Scholar] [CrossRef]
- Ilgü, H.; Jeckelmann, J.-M.; Gapsys, V.; Ucurum, Z.; de Groot, B.L.; Fotiadis, D. Insights into the molecular basis for substrate binding and specificity of the wild-type L-arginine/agmatine antiporter AdiC. Proc. Natl. Acad. Sci. USA 2016, 113, 10358–10363. [Google Scholar] [CrossRef]
- Harder, D.; Fotiadis, D. Measuring substrate binding and affinity of purified membrane transport proteins using the scintillation proximity assay. Nat. Protoc. 2012, 7, 1569–1578. [Google Scholar] [CrossRef]
- Aw, R.; Polizzi, K.M. Can too many copies spoil the broth? Microb. Cell Fact. 2013, 12, 128. [Google Scholar] [CrossRef]
- Liu, D.; Zeng, X.A.; Sun, D.W.; Han, Z. Disruption and protein release by ultrasonication of yeast cells. Innov. Food Sci. Emerg. Technol. 2013, 18, 132–137. [Google Scholar] [CrossRef]
- Bonetti, S.; Hirschi, S.; Bosshart, P.D. Expression, purification and crystallization of an SLC16 monocarboxylate transporter family homologue specific for l-lactate. Protein Expr. Purif. 2020, 165, 105484. [Google Scholar] [CrossRef]
- Jeckelmann, J.-M.; Harder, D.; Mari, S.A.; Meury, M.; Ucurum, Z.; Muller, D.J.; Erni, B.; Fotiadis, D. Structure and function of the glucose PTS transporter from Escherichia coli. J. Struct. Biol. 2011, 176, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef]
- Kotov, V.; Bartels, K.; Veith, K.; Josts, I.; Subhramanyam, U.K.T.; Gunther, C.; Labahn, J.; Marlovits, T.C.; Moraes, I.; Tidow, H.; et al. High-throughput stability screening for detergent-solubilized membrane proteins. Sci. Rep. 2019, 9, 10379. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Lee, Y.; Wiriyasermkul, P.; Tanaka, Y.; Takemoto, M.; Yamashita, K.; Nagamori, S.; Nishizawa, T.; Nureki, O. Consensus mutagenesis approach improves the thermal stability of system xc(-) transporter, xCT, and enables cryo-EM analyses. Protein Sci. 2020, 29, 2398–2407. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Kolmakova-Partensky, L.; Miller, C. A bacterial arginine-agmatine exchange transporter involved in extreme acid resistance. J. Biol. Chem. 2007, 282, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Casagrande, F.; Ratera, M.; Schenk, A.D.; Chami, M.; Valencia, E.; Lopez, J.M.; Torrents, D.; Engel, A.; Palacin, M.; Fotiadis, D. Projection structure of a member of the amino acid/polyamine/organocation transporter superfamily. J. Biol. Chem. 2008, 283, 33240–33248. [Google Scholar] [CrossRef] [PubMed]
| Purification Procedure | Yields 1 | |
|---|---|---|
| Membrane preparation | Membranes isolated | 7–9 g |
| Protein purification | Pellet after solubilization | 1.0–1.5 g |
| Protein amount after Ni-NTA elution | 13.2–14.8 mg | |
| Protein amount after Strep-Tag elution | 7.8–8.9 mg | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kantipudi, S.; Harder, D.; Bonetti, S.; Fotiadis, D.; Jeckelmann, J.-M. Pichia pastoris and the Recombinant Human Heterodimeric Amino Acid Transporter 4F2hc-LAT1: From Clone Selection to Pure Protein. Methods Protoc. 2021, 4, 51. https://doi.org/10.3390/mps4030051
Kantipudi S, Harder D, Bonetti S, Fotiadis D, Jeckelmann J-M. Pichia pastoris and the Recombinant Human Heterodimeric Amino Acid Transporter 4F2hc-LAT1: From Clone Selection to Pure Protein. Methods and Protocols. 2021; 4(3):51. https://doi.org/10.3390/mps4030051
Chicago/Turabian StyleKantipudi, Satish, Daniel Harder, Sara Bonetti, Dimitrios Fotiadis, and Jean-Marc Jeckelmann. 2021. "Pichia pastoris and the Recombinant Human Heterodimeric Amino Acid Transporter 4F2hc-LAT1: From Clone Selection to Pure Protein" Methods and Protocols 4, no. 3: 51. https://doi.org/10.3390/mps4030051
APA StyleKantipudi, S., Harder, D., Bonetti, S., Fotiadis, D., & Jeckelmann, J.-M. (2021). Pichia pastoris and the Recombinant Human Heterodimeric Amino Acid Transporter 4F2hc-LAT1: From Clone Selection to Pure Protein. Methods and Protocols, 4(3), 51. https://doi.org/10.3390/mps4030051






