Robust Normalization of Luciferase Reporter Data
Abstract
:1. Introduction
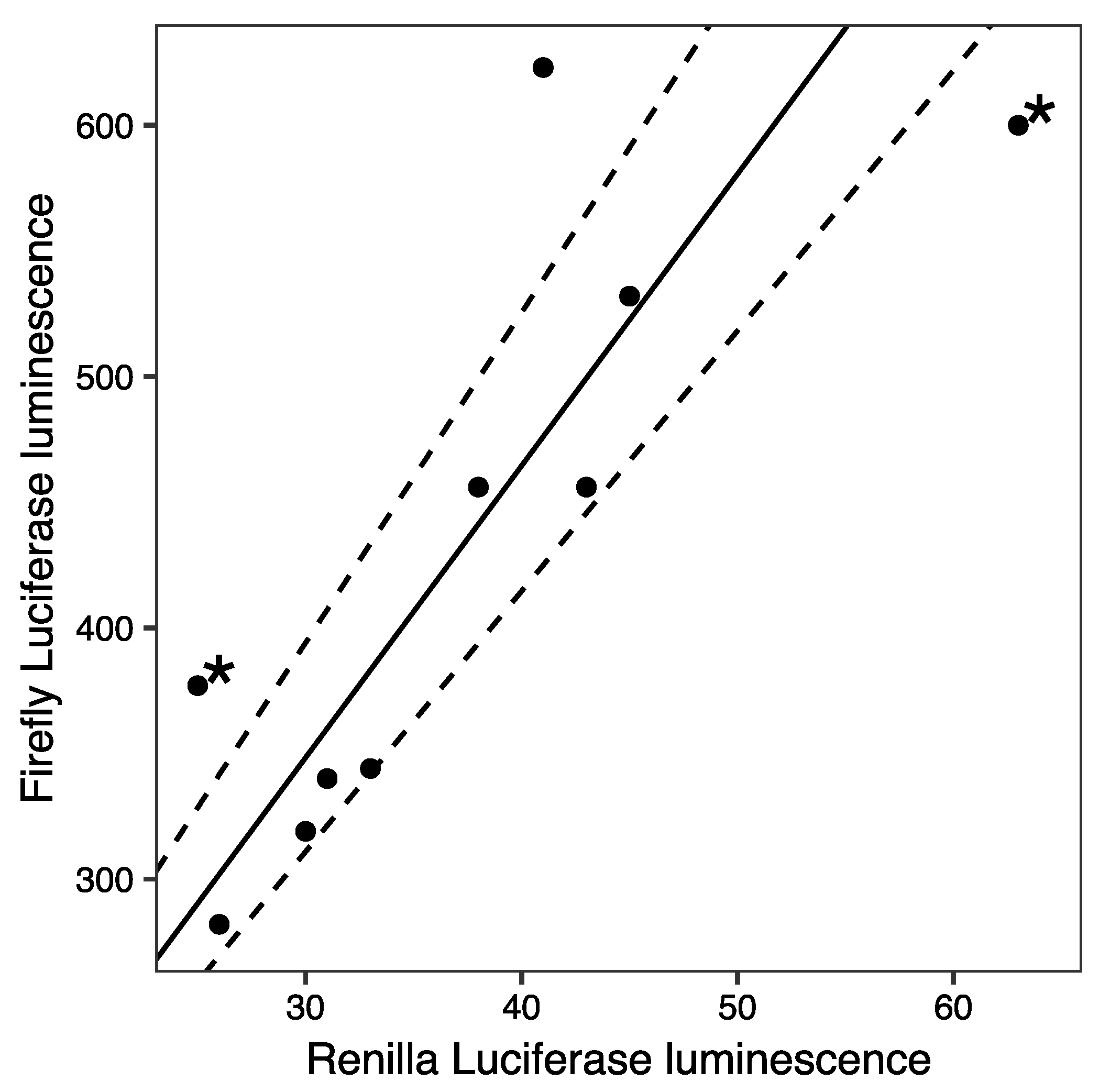
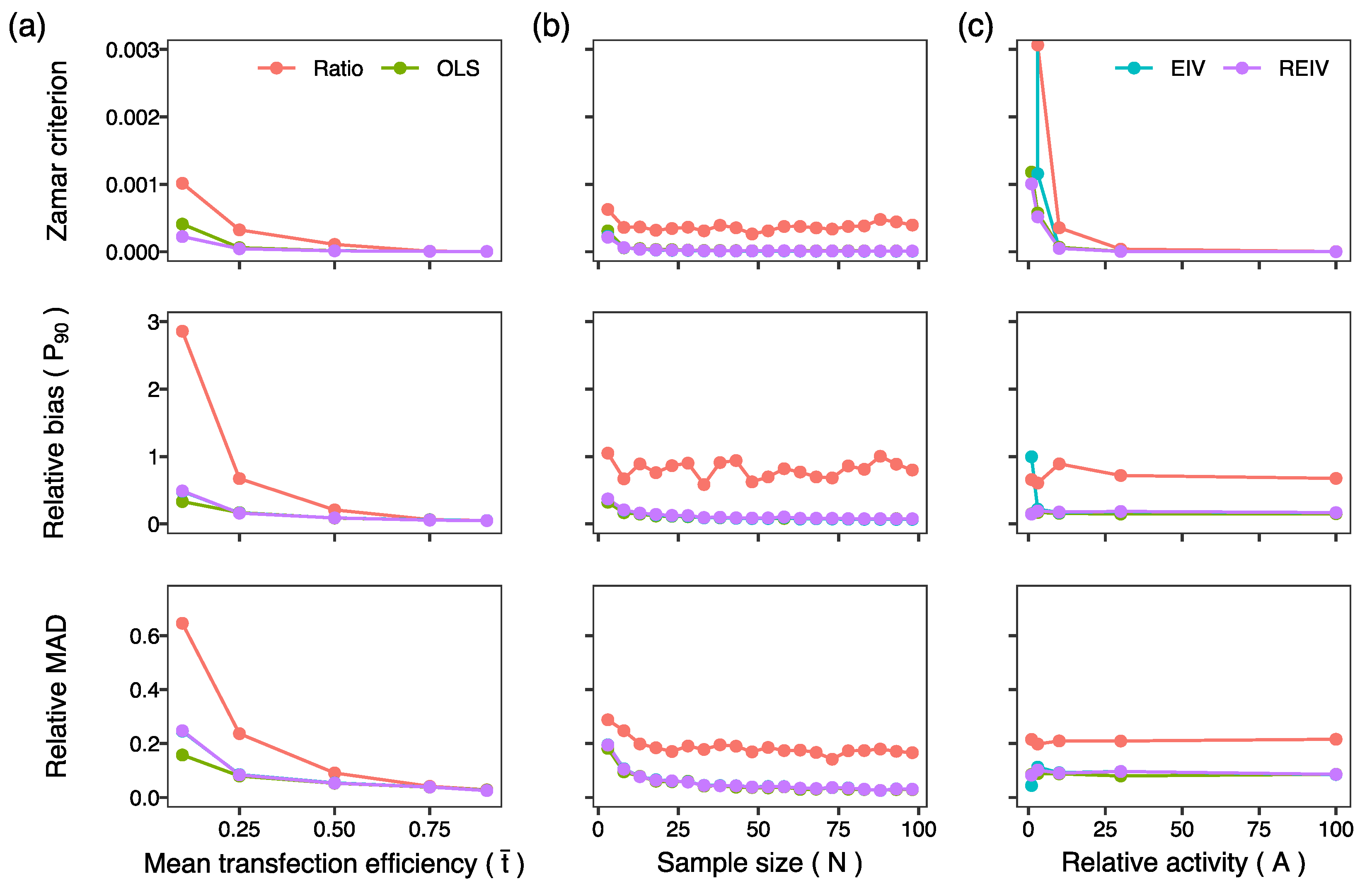
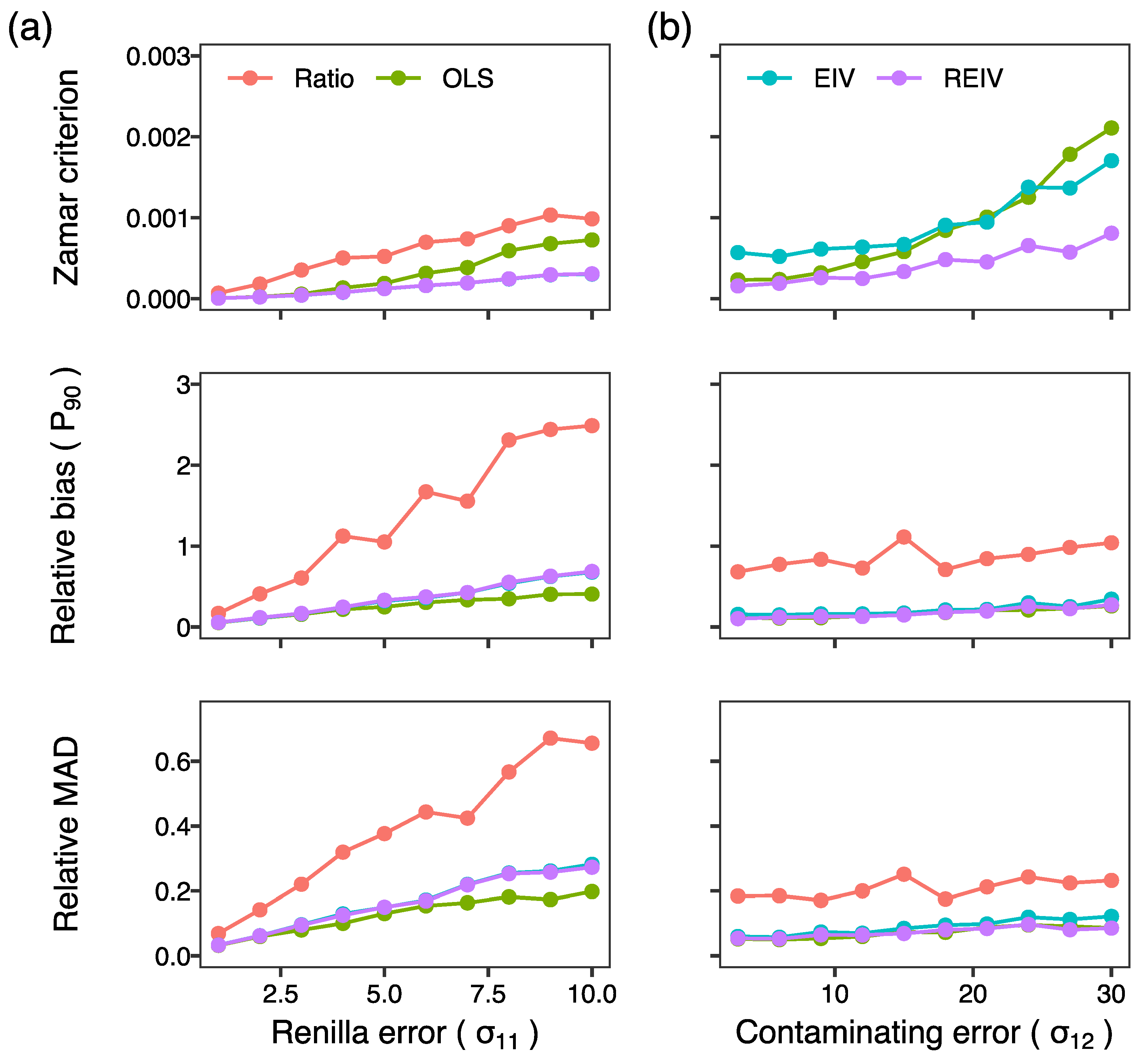
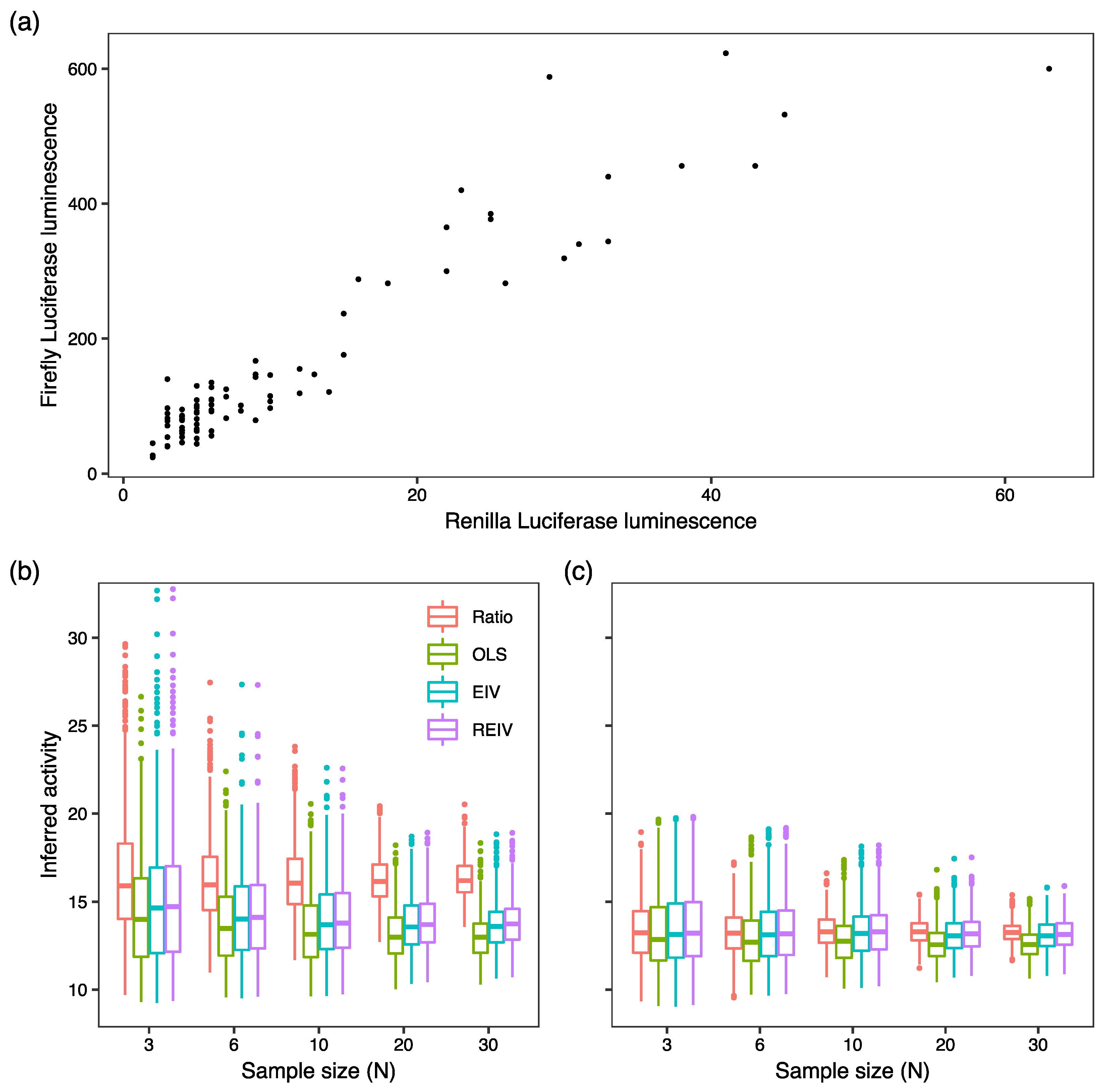
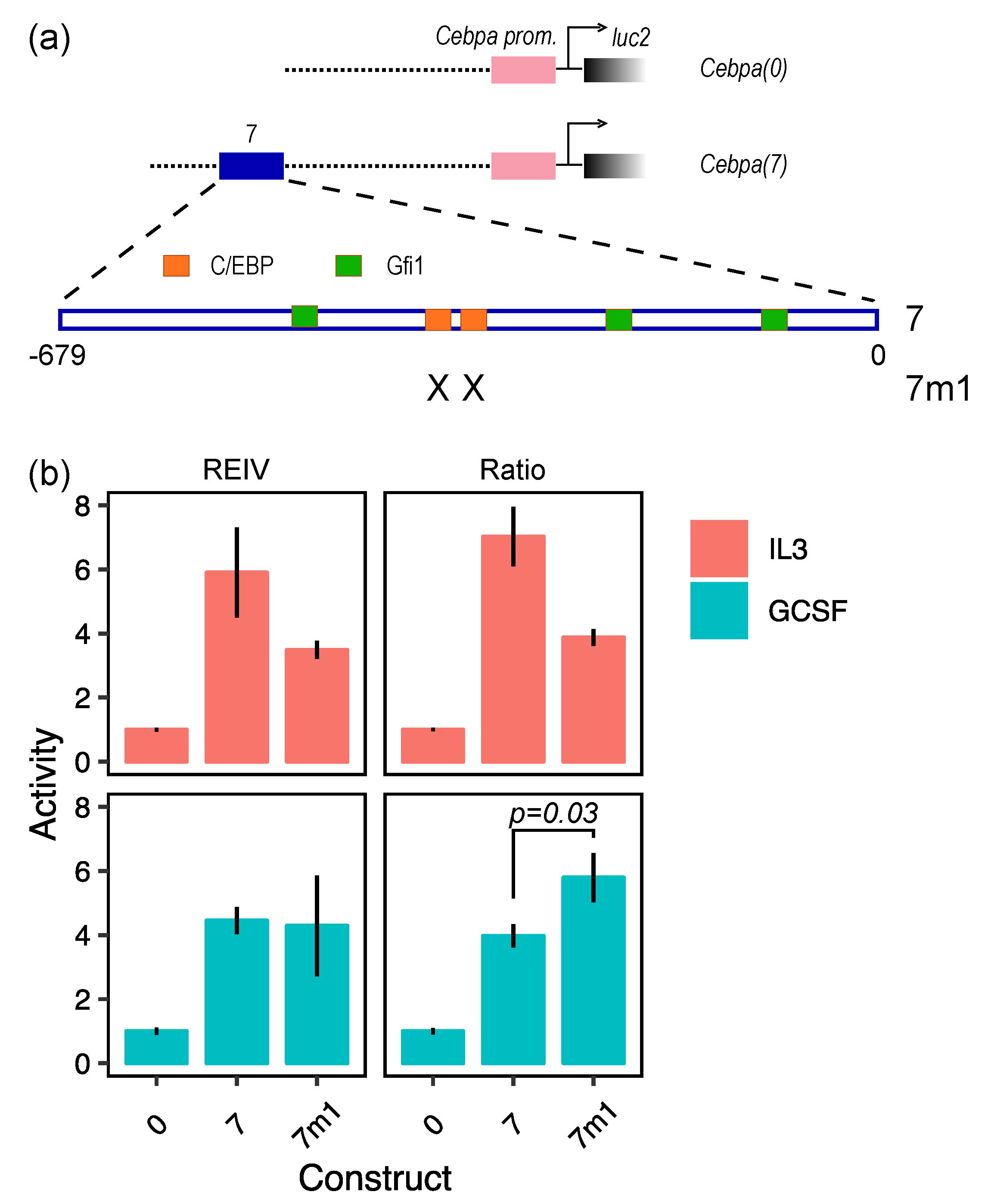
2. Results and Discussion
3. Materials and Methods
3.1. Cell Culture
3.2. Construction of the Luciferase Reporter Plasmid
3.3. Transfection and Luciferase Assays
3.4. Tested Normalization Methods
3.4.1. Ratio
3.4.2. Ordinary Least-Squares Regression
3.4.3. Errors-in-Variable Regression
3.4.4. Robust Errors-in-Variable Regression
3.5. Generation of Simulated Data
3.6. Procedure for Using REIV to Analyze Dual-Luciferase Reporter Data
3.6.1. Installing Required R Packages
- boot
- parallel
- nloptr
- ggplot2
- reshape2
- RColorBrewer
install.packages("<package_name>")3.6.2. Downloading REIV Code from GitHub and Loading It into R
- Download the code by visiting https://github.com/mlekkha/LUCNORM, clicking the Clone or download button and choosing Download ZIP.
- Uncompress the downloaded ZIP file.
- Set the working directory in R to the location of the downloaded code using Misc → Change Working Directory…
- Load the REIV analysis functions into the R workspace.source("lucAnalysis.R")source("robusteiv.R")
3.6.3. Input Data Format
3.6.4. Importing Input Data into the R Workspace and Preparing It
- In the R Console, read the file into a variable.data <- read.csv("<input_file_name>")
- (Optional) If any of the construct/condition names are numeric, they must be set as categorical variables. This may be accomplished as follows.data$<variable_name> <- as.factor(data$<variable_name>)
- For example:data$Construct <- as.factor(data$Construct)
3.6.5. Normalizing and Saving Results
- Once the data have been saved in a variable, REIV can be used to compute normalized Luciferase activity and its confidence interval for each combination of construct and condition.norm_activity <- calcSlopesCIs(<data_variable_name>,alpha = <alpha_value>,regmethod = robusteiv,cim = "boot_positive_ci",ignore=c("Luc", "Ren"))Here, alpha is the significance threshold. For example, setting alpha to will compute 95% confidence intervals. regmethod is the normalization method. Use regmethod = eiv for EIV normalization. cim is the method used for computing the confidence intervals. Use cim = "gleser_ci" for EIV normalization. ignore specifies which columns/variables are not conditions. If there are other columns in the CSV file that are not conditions, such as annotations, they should be included in the ignore vector.Example usage:norm_activity <- calcSlopesCIs(data,alpha=0.05,regmethod=robusteiv,cim="boot_positive_ci",ignore=c("Luc", "Ren"))The output of calcSlopesCIs is a data frame with columns containing the normalized Luciferase activity (slope) and the lower and upper confidence intervals (ci_lower and ci_upper). The values may be inspected with the print command.print(norm_activity)
- The normalized activities may be saved to a CSV file for further analysis or visualization using the write.csv function.write.csv(<variable_name>, "<output_file_name>")For example:write.csv(norm_activity, "normalized_activities.csv")
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PUER | PU.1 Estrogen Receptor |
| IL3 | Interleukin 3 |
| GCSF | Granulocyte colony stimulating factor |
| OLS | Ordinary least-squares |
| EIV | Errors-in-variables |
| REIV | Robust errors-in-variables |
References
- Arnold, C.D.; Gerlach, D.; Stelzer, C.; Boryn, L.M.; Rath, M.; Stark, A. Genome-Wide Quantitative Enhancer Activity Maps Identified by STARR-seq. Science 2013. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Laslo, P.; Spooner, C.J.; Warmflash, A.; Lancki, D.W.; Lee, H.J.; Sciammas, R.; Gantner, B.N.; Dinner, A.R.; Singh, H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 2006, 126, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Repele, A.; Krueger, S.; Bhattacharyya, T.; Tuineau, M.Y.; Manu. The regulatory control of Cebpa enhancers and silencers in the myeloid and red-blood cell lineages. PLoS ONE 2019, 14, e0217580. [Google Scholar] [CrossRef] [PubMed]
- Stratowa, C.; Himmler, A.; Czernilofsky, A.P. Use of a luciferase reporter system for characterizing G-protein-linked receptors. Curr. Opin. Biotechnol. 1995, 6, 574–581. [Google Scholar] [CrossRef]
- Savkur, R.S.; Bramlett, K.S.; Stayrook, K.R.; Nagpal, S.; Burris, T.P. Coactivation of the human vitamin D receptor by the peroxisome proliferator-activated receptor gamma coactivator-1 alpha. Mol. Pharmacol. 2005, 68, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Sanada, M.; Kato, I.; Sato, Y.; Takita, J.; Takeuchi, K.; Niwa, A.; Chen, Y.; Nakazaki, K.; Nomoto, J.; et al. Frequent inactivation of A20 in B-cell lymphomas. Nature 2009, 459, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.L.; Dinman, J.D. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004, 32, e160. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Wood, K.V. Bioluminescent assays for high-throughput screening. Assay Drug Dev. Technol. 2007, 5, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Minkovsky, A.; Sahakyan, A.; Bonora, G.; Damoiseaux, R.; Dimitrova, E.; Rubbi, L.; Pellegrini, M.; Radu, C.G.; Plath, K. A high-throughput screen of inactive X chromosome reactivation identifies the enhancement of DNA demethylation by 5-aza-2′-dC upon inhibition of ribonucleotide reductase. Epigenetics Chromatin 2015, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Smale, S.T. Luciferase assay. Cold Spring Harb. Protoc. 2010. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, M.S.; Brownlee, G.G. Cis-acting elements and transcription factors involved in the promoter activity of the human factor VIII gene. J. Biol. Chem. 1995, 270, 11828–11838. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.C.; DeKoter, R.P.; Lee, H.J.; Smith, E.D.; Lancki, D.W.; Gurish, M.F.; Friend, D.S.; Stevens, R.L.; Anastasi, J.; Singh, H. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity 2002, 17, 665–676. [Google Scholar] [CrossRef]
- Dahl, R.; Walsh, J.C.; Lancki, D.; Laslo, P.; Iyer, S.R.; Singh, H.; Simon, M.C. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat. Immunol. 2003, 4, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- Bertolino, E.; Reinitz, J.; Manu. The analysis of novel distal Cebpa enhancers and silencers using a transcriptional model reveals the complex regulatory logic of hematopoietic lineage specification. Dev. Biol. 2016, 413, 128–144. [Google Scholar] [CrossRef] [PubMed]
- Casella, G.; Berger, R.L. Statistical Inference, 2nd ed.; Duxbury Press: Pacific Grove, CA, USA, 2001. [Google Scholar]
- Zamar, R.H. Robust Estimation in the Errors-in-Variables Model. Biometrika 1989, 76, 149–160. [Google Scholar] [CrossRef]
- Legraverend, C.; Antonson, P.; Flodby, P.; Xanthopoulos, K.G. High level activity of the mouse CCAAT/enhancer binding protein (C/EBP alpha) gene promoter involves autoregulation and several ubiquitous transcription factors. Nucleic Acids Res. 1993, 21, 1735–1742. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
| 0.1 | 2 | 18 |
| 0.25 | 2 | 6 |
| 0.5 | 2 | 2 |
| 0.75 | 6 | 2 |
| 0.9 | 18 | 2 |
| Luc | Ren | Construct | Condition 1 | Condition 2 | ⋯ |
|---|---|---|---|---|---|
| Luc luminescence | Ren luminescence | Name | Name | Name | ⋯ |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Repele, A.; Manu. Robust Normalization of Luciferase Reporter Data. Methods Protoc. 2019, 2, 62. https://doi.org/10.3390/mps2030062
Repele A, Manu. Robust Normalization of Luciferase Reporter Data. Methods and Protocols. 2019; 2(3):62. https://doi.org/10.3390/mps2030062
Chicago/Turabian StyleRepele, Andrea, and Manu. 2019. "Robust Normalization of Luciferase Reporter Data" Methods and Protocols 2, no. 3: 62. https://doi.org/10.3390/mps2030062
APA StyleRepele, A., & Manu. (2019). Robust Normalization of Luciferase Reporter Data. Methods and Protocols, 2(3), 62. https://doi.org/10.3390/mps2030062





