Novel ‘Bacteriospray’ Method Facilitates the Functional Screening of Metagenomic Libraries for Antimicrobial Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmid Growth Conditions
2.2. Library Construction Strategies
2.3. Derivation of a Chloramphenicol Resistant Strain
2.4. Top Agar Overlay Method
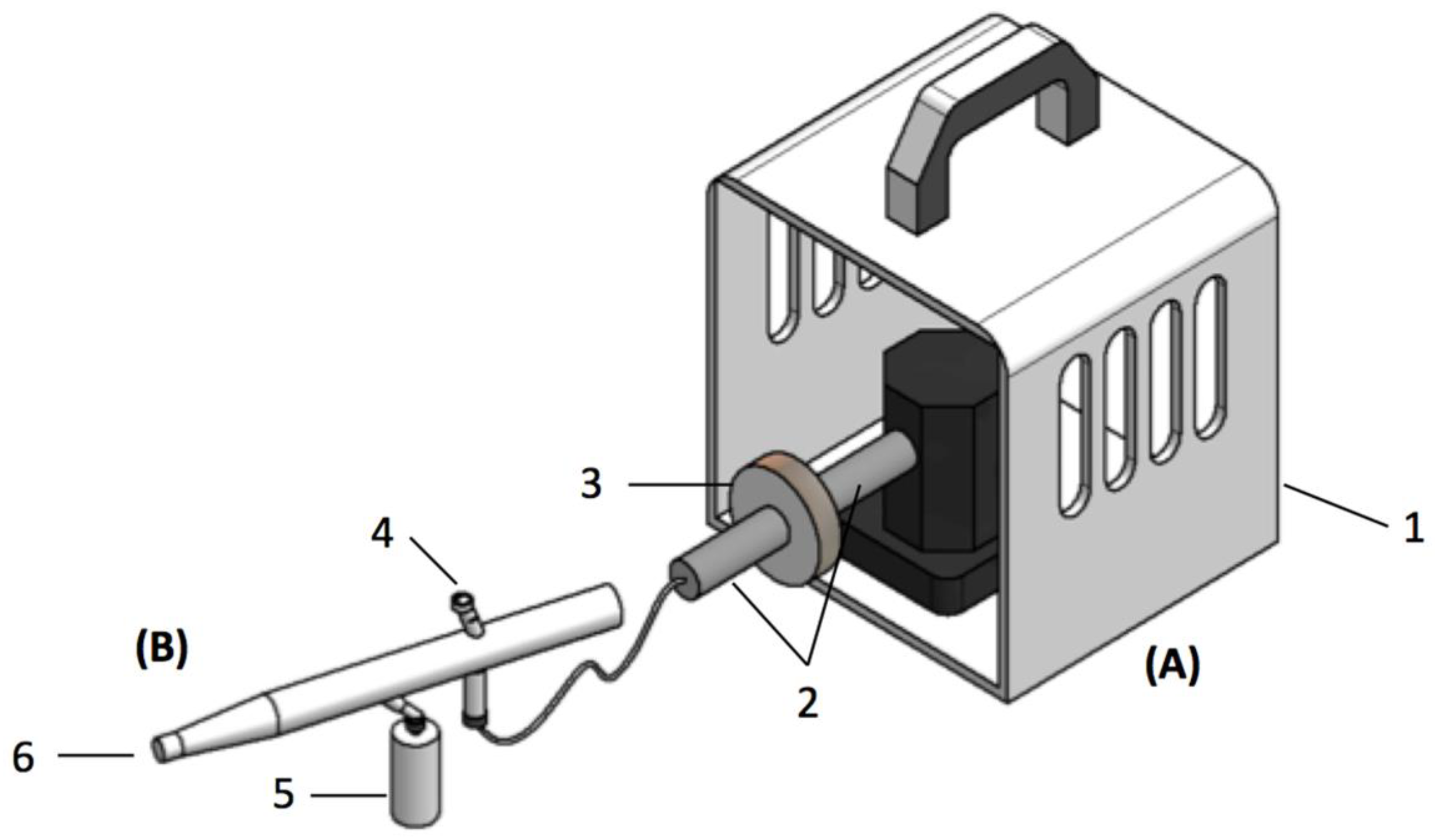
2.5. Bacteriospray Screening Method
2.6. Growth Inhibition Zone Confirmation, DNA Sequencing and Insert Analysis
2.7. Statistical Analysis
3. Results
3.1. Transformation Efficiency Optimization
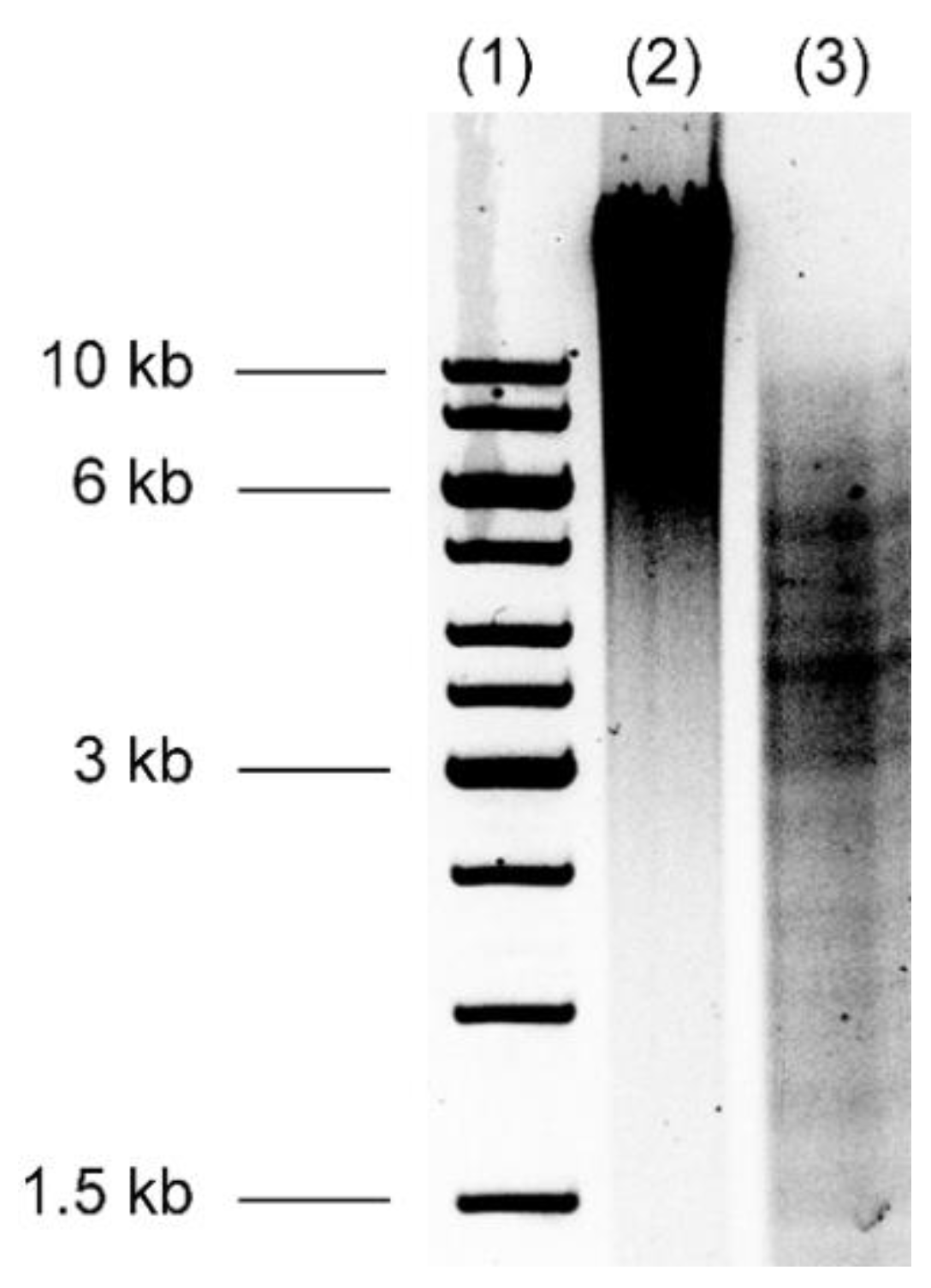
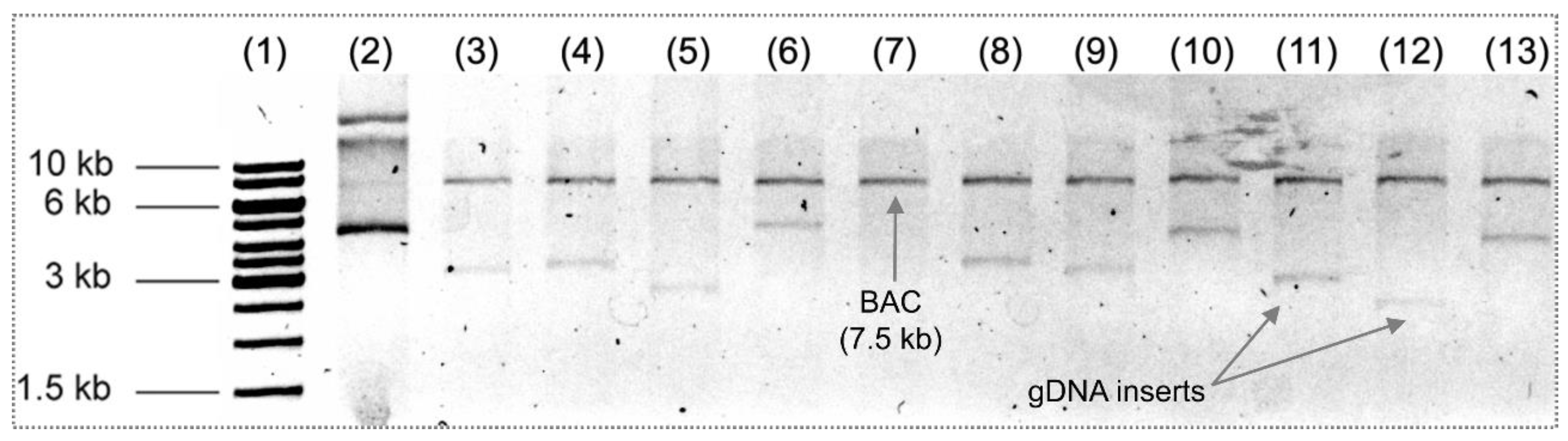
3.2. Burkholderia thailandensis Library Construction
3.3. Top Agar Overlay vs Bacteriospray Assay for the Functional Screening of Metagenomic Libraries
3.4. Sequence Analysis of Inserted Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu, H.; Swierstra, J.; Wu, C.; Girard, G.; Choi, Y.H.; van Wamel, W.; Sandiford, S.K.; van Wezel, G.P. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology 2014, 160, 1714–1725. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. (In IRIS); World Health Organization: Geneva, Switzerland, 2014; p. 232. [Google Scholar]
- Blaskovich, M.A.; Butler, M.S.; Cooper, M.A. Polishing the tarnished silver bullet: The quest for new antibiotics. Essays Biochem. 2017, 61, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Opportunities for natural products in 21st century antibiotic discovery. Nat. Prod. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef]
- Wang, G.Y.; Graziani, E.; Waters, B.; Pan, W.; Li, X.; McDermott, J.; Meurer, G.; Saxena, G.; Andersen, R.J.; Davies, J. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2000, 2, 2401–2404. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.Y.; Brady, S.F. Discovery of indolotryptoline antiproliferative agents by homology-guided metagenomic screening. Proc. Natl. Acad. Sci. USA 2013, 110, 2478–2483. [Google Scholar] [CrossRef]
- Lim, H.K.; Chung, E.J.; Kim, J.C.; Choi, G.J.; Jang, K.S.; Chung, Y.R.; Cho, K.Y.; Lee, S.W. Characterization of a forest soil metagenome clone that confers indirubin and indigo production on Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 7768–7777. [Google Scholar] [CrossRef]
- Rajendhran, J.; Gunasekaran, P. Strategies for accessing soil metagenome for desired applications. Biotechnol. Adv. 2008, 26, 576–590. [Google Scholar] [CrossRef]
- Parachin, N.S.; Schelin, J.; Norling, B.; Radstrom, P.; Gorwa-Grauslund, M.F. Flotation as a tool for indirect DNA extraction from soil. Appl. Microbiol. Biotechnol. 2010, 87, 1927–1933. [Google Scholar] [CrossRef] [PubMed]
- Delmont, T.O.; Robe, P.; Clark, I.; Simonet, P.; Vogel, T.M. Metagenomic comparison of direct and indirect soil DNA extraction approaches. J. Microbiol. Methods 2011, 86, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Ferrand, J.; Patron, K.; Legrand-Frossi, C.; Frippiat, J.P.; Merlin, C.; Alauzet, C.; Lozniewski, A. Comparison of seven methods for extraction of bacterial DNA from fecal and cecal samples of mice. J. Microbiol. Methods 2014, 105, 180–185. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, K.K.; Kuhad, R.C. An efficient and economical method for extraction of DNA amenable to biotechnological manipulations, from diverse soils and sediments. J. Appl. Microbiol. 2014, 116, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat. Protoc. 2007, 2, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Daniel, R. Construction of small-insert and large-insert metagenomic libraries. Methods Mol. Biol. 2010, 668, 39–50. [Google Scholar] [CrossRef]
- de Castro, A.P.; Quirino, B.F.; Allen, H.; Williamson, L.L.; Handelsman, J.; Kruger, R.H. Construction and validation of two metagenomic DNA libraries from Cerrado soil with high clay content. Biotechnol. Lett 2011, 33, 2169–2175. [Google Scholar] [CrossRef]
- Kakirde, K.S.; Parsley, L.C.; Liles, M.R. Size Does Matter: Application-driven Approaches for Soil Metagenomics. Soil Biol. Biochem. 2010, 42, 1911–1923. [Google Scholar] [CrossRef]
- Rondon, M.R.; August, P.R.; Bettermann, A.D.; Brady, S.F.; Grossman, T.H.; Liles, M.R.; Loiacono, K.A.; Lynch, B.A.; MacNeil, I.A.; Minor, C. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000, 66, 2541–2547. [Google Scholar] [CrossRef]
- Craig, J.W.; Chang, F.Y.; Kim, J.H.; Obiajulu, S.C.; Brady, S.F. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl. Environ. Microbiol. 2010, 76, 1633–1641. [Google Scholar] [CrossRef]
- Kim, U.-J.; Birren, B.W.; Slepak, T.; Mancino, V.; Boysen, C.; Kang, H.-L.; Simon, M.I.; Shizuya, H. Construction and characterization of a human bacterial artificial chromosome library. Genomics 1996, 34, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Hunt, D.; Sandham, H. Improved agar gradient-plate technique. Appl. Microbiol. 1969, 17, 329. [Google Scholar] [PubMed]
- Brady, S.F.; Chao, C.J.; Handelsman, J.; Clardy, J. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 2001, 3, 1981–1984. [Google Scholar] [CrossRef] [PubMed]
- MacNeil, I.; Tiong, C.; Minor, C.; August, P.; Grossman, T.; Loiacono, K.; Lynch, B.; Phillips, T.; Narula, S.; Sundaramoorthi, R. Expression and isolation of antimicrobial small molecules from soil DNA libraries. J. Mol. Microbiol. Biotechnol. 2001, 3, 301–308. [Google Scholar] [PubMed]
- Gillespie, D.E.; Brady, S.F.; Bettermann, A.D.; Cianciotto, N.P.; Liles, M.R.; Rondon, M.R.; Clardy, J.; Goodman, R.M.; Handelsman, J. Isolation of Antibiotics Turbomycin A and B from a Metagenomic Library of Soil Microbial DNA. Appl. Environ. Microbiol. 2002, 68, 4301–4306. [Google Scholar] [CrossRef]
- Seeburg, P.H.; Shine, J.; Martial, J.A.; Baxter, J.D.; Goodman, H.M. Nucleotide sequence and amplification in bacteria of structural gene for rat growth hormone. Nature 1977, 270, 486. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Romantsov, T.; Engel, K.; Doxey, A.C.; Rose, D.R.; Neufeld, J.D.; Charles, T.C. Functional metagenomics reveals novel beta-galactosidases not predictable from gene sequences. PLoS ONE 2017, 12, e0172545. [Google Scholar] [CrossRef]
- Mao, D.; Bushin, L.B.; Moon, K.; Wu, Y.; Seyedsayamdost, M.R. Discovery of scmR as a global regulator of secondary metabolism and virulence in Burkholderia thailandensis E264. Proc. Natl. Acad. Sci. USA 2017, 114, E2920–E2928. [Google Scholar] [CrossRef]
- Rondon, M.R.; Raffel, S.J.; Goodman, R.M.; Handelsman, J. Toward functional genomics in bacteria: Analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc. Natl. Acad. Sci. 1999, 96, 6451–6455. [Google Scholar] [CrossRef]
- Sabree, Z.L.; Rondon, M.R.; Handelsman, J. Metagenomics. In Encyclopedia of Microbiology (Third Edition); Schaechter, M., Ed.; Academic Press: Oxford, UK, 2009; pp. 622–632. [Google Scholar] [CrossRef]
- Lussier, F.X.; Chambenoit, O.; Cote, A.; Hupe, J.F.; Denis, F.; Juteau, P.; Beaudet, R.; Shareck, F. Construction and functional screening of a metagenomic library using a T7 RNA polymerase-based expression cosmid vector. J. Ind. Microbiol. Biotechnol. 2011, 38, 1321–1328. [Google Scholar] [CrossRef]
- Sosio, M.; Giusino, F.; Cappellano, C.; Bossi, E.; Puglia, A.M.; Donadio, S. Artificial chromosomes for antibiotic-producing actinomycetes. Nat. Biotechnol. 2000, 18, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Biggins, J.B.; Liu, X.; Feng, Z.; Brady, S.F. Metabolites from the induced expression of cryptic single operons found in the genome of Burkholderia pseudomallei. J. Am. Chem. Soc. 2011, 133, 1638–1641. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cheng, Y.Q. Genome-guided discovery of diverse natural products from Burkholderia sp. J. Ind. Microbiol. Biotechnol. 2014, 41, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Sultana, A.; Kallio, P.; Jansson, A.; Wang, J.S.; Niemi, J.; Mäntsälä, P.; Schneider, G. Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation. EMBO J. 2004, 23, 1911–1921. [Google Scholar] [CrossRef] [PubMed]
- de Jong, A.; van Hijum, S.A.; Bijlsma, J.J.; Kok, J.; Kuipers, O.P. BAGEL: A web-based bacteriocin genome mining tool. Nucleic Acids Res. 2006, 34, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Raguse, M.; Fiebrandt, M.; Stapelmann, K.; Madela, K.; Laue, M.; Lackmann, J.W.; Thwaite, J.E.; Setlow, P.; Awakowicz, P.; Moeller, R. Improvement of Biological Indicators by Uniformly Distributing Bacillus subtilis Spores in Monolayers To Evaluate Enhanced Spore Decontamination Technologies. Appl. Environ. Microbiol. 2016, 82, 2031–2038. [Google Scholar] [CrossRef]
- Paton, S.; Thompson, K.A.; Parks, S.R.; Bennett, A.M. Reaerosolization of Spores from Flooring Surfaces To Assess the Risk of Dissemination and Transmission of Infections. Appl. Environ. Microbiol. 2015, 81, 4914–4919. [Google Scholar] [CrossRef]
- Levy, C.; Bornard, I.; Carlin, F. Deposition of Bacillus subtilis spores using an airbrush-spray or spots to study surface decontamination by pulsed light. J. Microbiol. Methods 2011, 84, 223–227. [Google Scholar] [CrossRef]




| Clone Locus Tags | Similar Protein (Accession Number) |
| pBeloBAC11 B1 | |
| BTH_II0226 | Snoal-like protein (3H3H A) |
| BTH_II0227 | Conserved hypothetical protein (ABC35626.1) |
| pBeloBAC11 B2 | |
| BTH_II0243 | LysR family transcriptional regulator (WP_009894988.1) |
| BTH_II0244 | LysR family transcriptional regulator (WP_009894990.1) |
| BTH_II0245 | dsdA D-serindeshydratase (Q2T8Q2.1) |
| BTH_II0246 | LysR family transcriptional regulator (WP_011400849.1) |
| pBeloBAC11 B3 & B4 | |
| BTH_II0203 | Carbohydrate diacid regulator (ABC35448.1) |
| BTH_II0204 | Peptide synthase, putative (ABC34379.1) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brahami, A.; Castonguay, A.; Déziel, É. Novel ‘Bacteriospray’ Method Facilitates the Functional Screening of Metagenomic Libraries for Antimicrobial Activity. Methods Protoc. 2019, 2, 4. https://doi.org/10.3390/mps2010004
Brahami A, Castonguay A, Déziel É. Novel ‘Bacteriospray’ Method Facilitates the Functional Screening of Metagenomic Libraries for Antimicrobial Activity. Methods and Protocols. 2019; 2(1):4. https://doi.org/10.3390/mps2010004
Chicago/Turabian StyleBrahami, Anissa, Annie Castonguay, and Éric Déziel. 2019. "Novel ‘Bacteriospray’ Method Facilitates the Functional Screening of Metagenomic Libraries for Antimicrobial Activity" Methods and Protocols 2, no. 1: 4. https://doi.org/10.3390/mps2010004
APA StyleBrahami, A., Castonguay, A., & Déziel, É. (2019). Novel ‘Bacteriospray’ Method Facilitates the Functional Screening of Metagenomic Libraries for Antimicrobial Activity. Methods and Protocols, 2(1), 4. https://doi.org/10.3390/mps2010004






